Graphical User Interface¶
Start the Graphical User Interface¶
In a terminal, enter to following:
$ source activate py27cmp-gui
or:
$ conda activate py27cmp-gui
Please see Section Installation for more details about installation.
After activation of the conda environment, start the graphical user interface called Connectome Mapper 3 BIDS App Manager
$ cmpbidsappmanager
Load a BIDS dataset¶
The Connectome Mapper 3 BIDS App Manager allows you to:
- load a BIDS dataset stored locally
You only have to select the root directory of your valid BIDS dataset (see note below)
- create a new datalad/BIDS dataset locally from an existing local or remote datalad/BIDS dataset (This is a feature under development)
Select the mode “Install a Datalad/BIDS dataset”.
If ssh connection is used, make sure to enable the “install via ssh” and to provide all connection details (IP address / Remote host name, remote user, remote password)
Note
The input dataset MUST be a valid BIDS structured dataset and must include at least one T1w or MPRAGE structural image. We highly recommend that you validate your dataset with the free, online BIDS Validator.
Pipeline stage configuration¶
Start the Configurator Window¶
- From the main window, click on the left button to start the Configurator Window.
- The window of the Connectome Mapper BIDS App Configurator will appear, which will assist you note only in configuring the pipeline stages (each pipeline has a tab panel), but also in creating appropriate configuration files which could be used outside the Graphical User Interface.
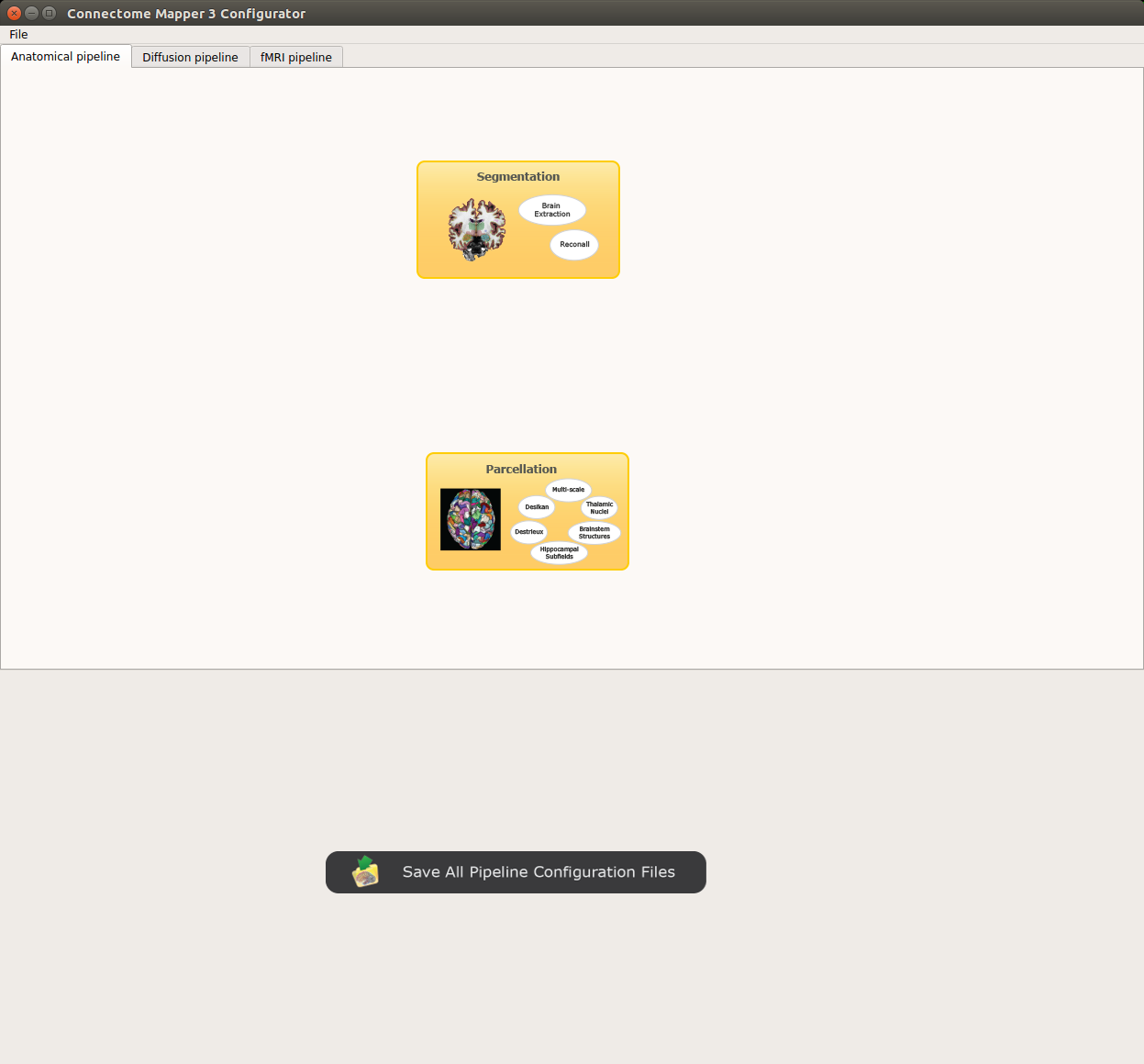
Configurator Window of the Connectome Mapper
The outputs depend on the chosen parameters.
Anatomical pipeline stages¶
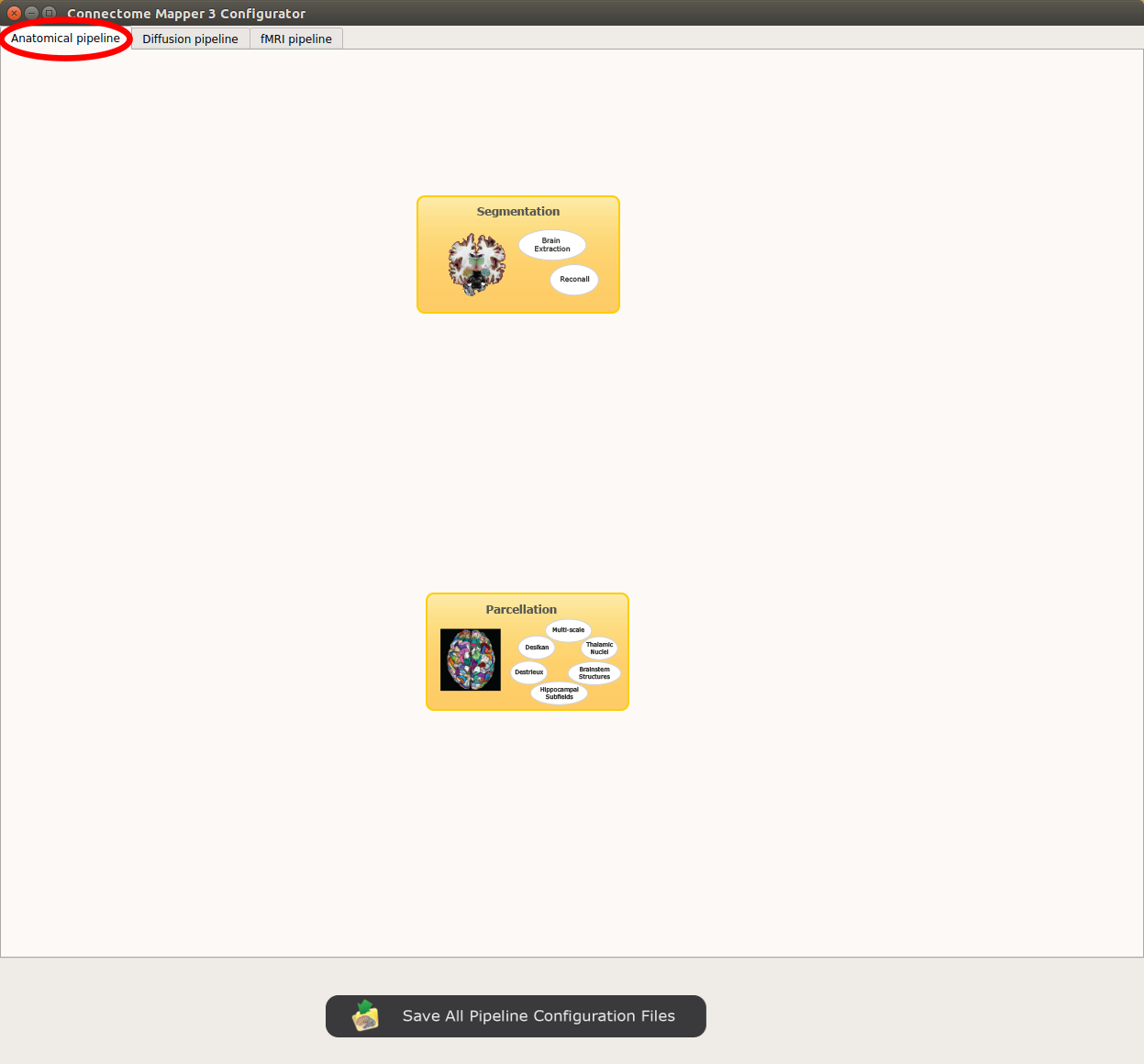
Panel for configuration of anatomical pipeline stages
Segmentation¶
Performs tissue segmentation using Freesurfer or custom segmentation.
Freesurfer
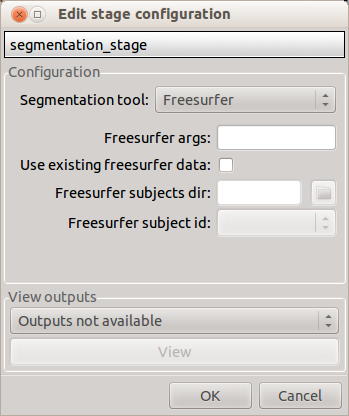
- Freesurfer args: used to specify Freesurfer processing options
- Use existing freesurfer data: Check this box if you have already Freesurfer output data available
Custom segmentation
Warning
Not fully tested. Development and testing in progress.
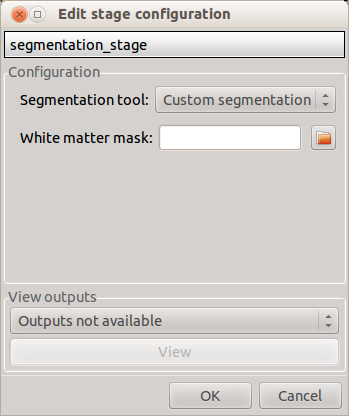
- White matter mask: select the file containing your white matter binary mask
Parcellation¶
Generates the Native Freesurfer or Lausanne2008/Lausanne2018 parcellation from Freesurfer data, or takes a custom parcellation atlas.
Parcellation scheme
NativeFreesurfer:
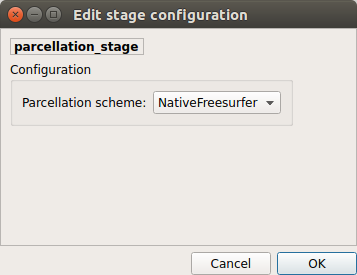
Atlas composed of 83 regions from the Freesurfer aparc+aseg file
Lausanne2008:
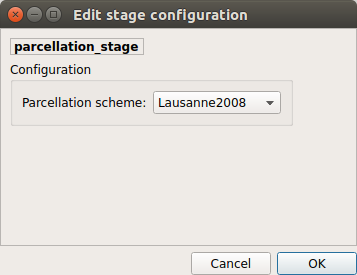
Multi-resolution atlas
Lausanne2018:
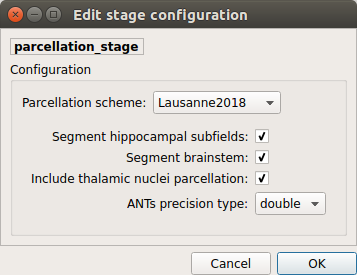
Lausanne 2008 atlas extended with 7 thalamic nuclei, 12 hippocampal subfields, and 4 brainstem sub-structure per hemisphere
Custom:
Warning
Not fully tested. Development and testing in progress.
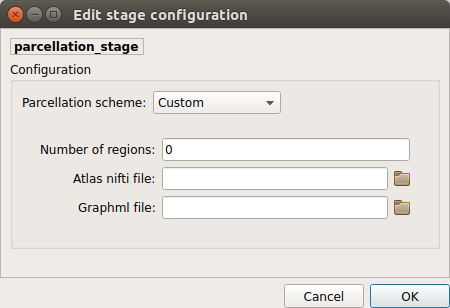
Custom atlas. Specify the atlas name, the number of regions, the nifti file and a corresponding graphml file. The Graphml file must contain at least a “dn_correspondence_id” field for each node. This field should contain the region’s label in the nifti file.
Diffusion pipeline stages¶
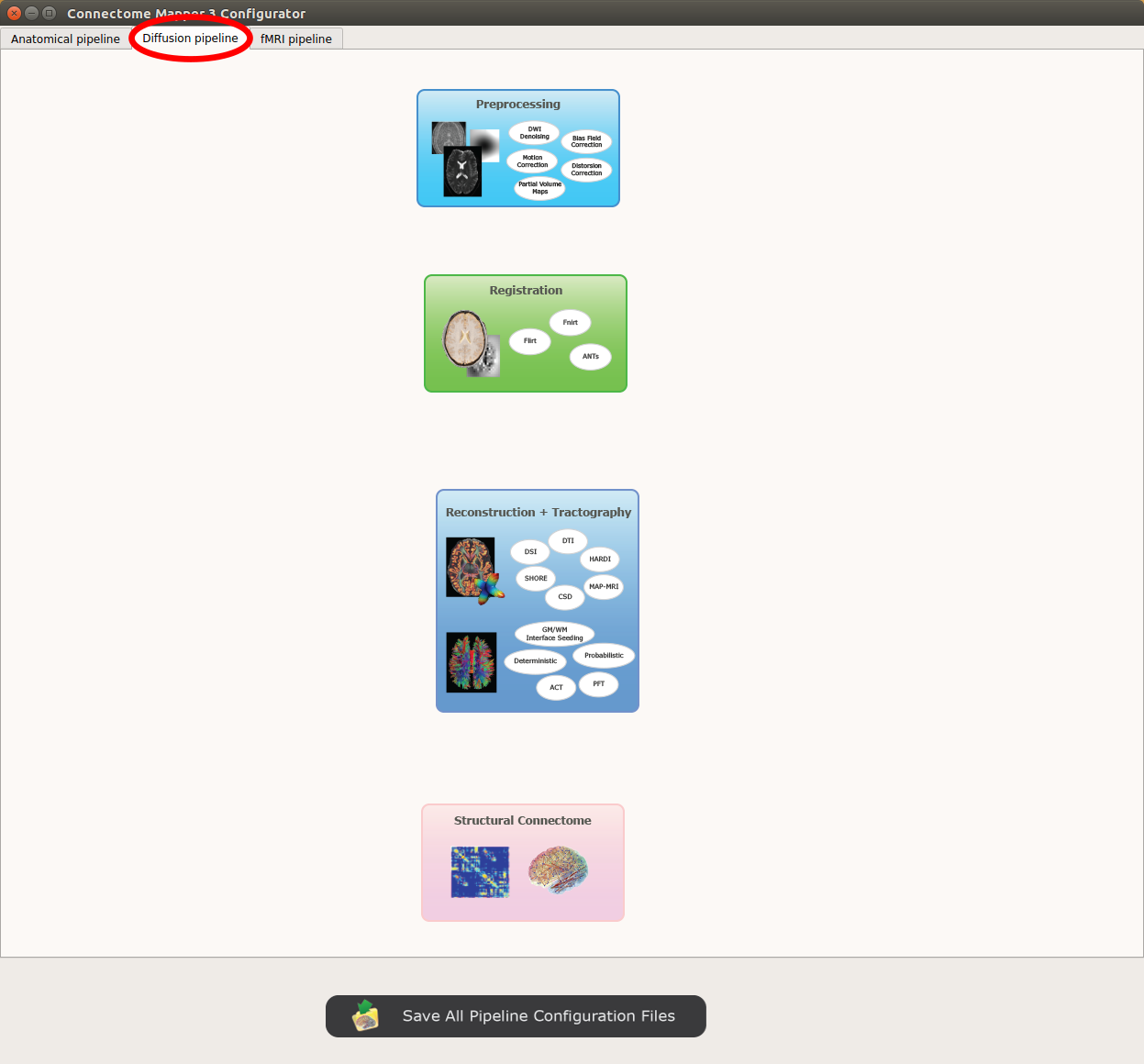
Panel for configuration of diffusion pipeline stages
Preprocessing¶
Preprocessing includes denoising, bias field correction, motion and eddy current correction for diffusion data.
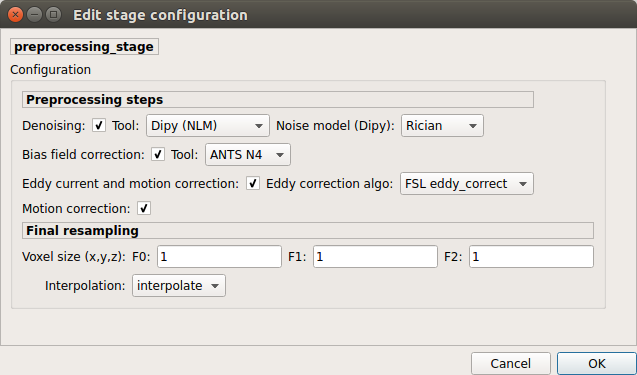
Denoising
Remove noise from diffusion images using (1) MRtrix3 MP-PCA method or (2) Dipy Non-Local Mean (NLM) denoising with Gaussian or Rician noise models
Bias field correction
Remove intensity inhomogeneities due to the magnetic resonnace bias field using (1) MRtrix3 N4 bias field correction or (2) the bias field correction provided by FSL FAST.
Motion correction
Aligns diffusion volumes to the b0 volume using FSL’s MCFLIRT.
Note
For hemi-sphere DSI aquisitions, warning outputs will be displayed in the console when processing empty volumes.
Eddy current correction
Corrects for eddy current distortions using FSL’s Eddy correct tool.
Resampling
Resample morphological and diffusion data to F0 x F1 x F2 mm^3
Registration¶
Registration mode
FSL (Linear):
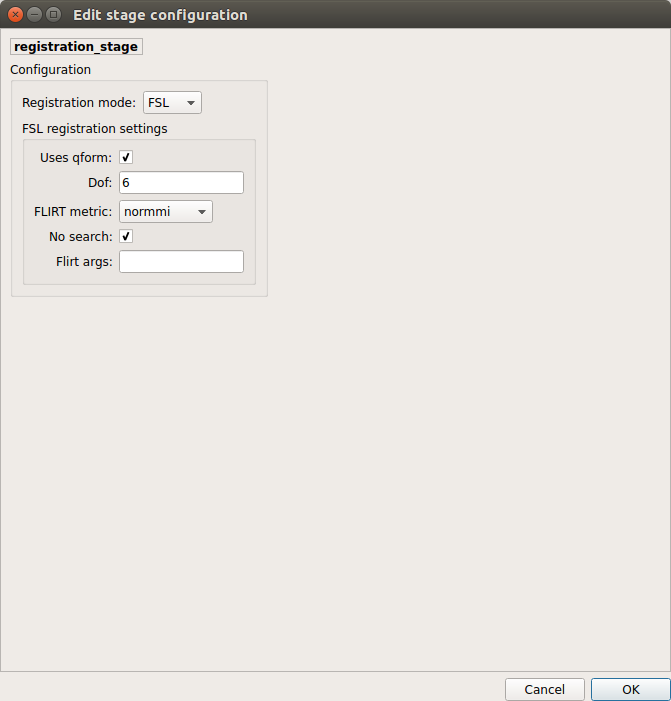
Perform linear registration from T1 to diffusion b0 using FSL’s flirt.
BBregister (FS):
Perform linear registration using Freesurfer BBregister tool.
Non-linear (ANTS):
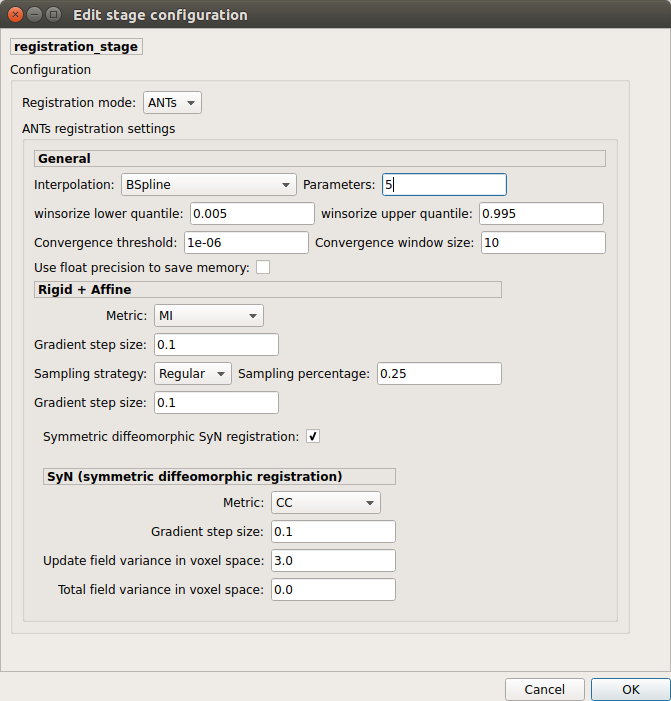
Perform symmetric diffeomorphic SyN registration from T1 to b0
Diffusion reconstruction and tractography¶
Perform diffusion reconstruction and local deterministic or probabilistic tractography based on several tools. ROI dilation is required to map brain connections when the tracking only operates in the white matter.
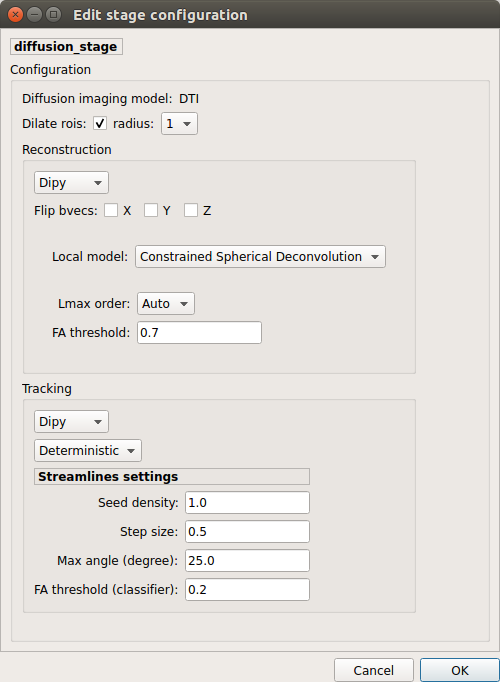
Diffusion stage configuration window
Reconstruction tool
Dipy: perform SHORE, tensor, CSD and MAP-MRI reconstruction.
SHORE:
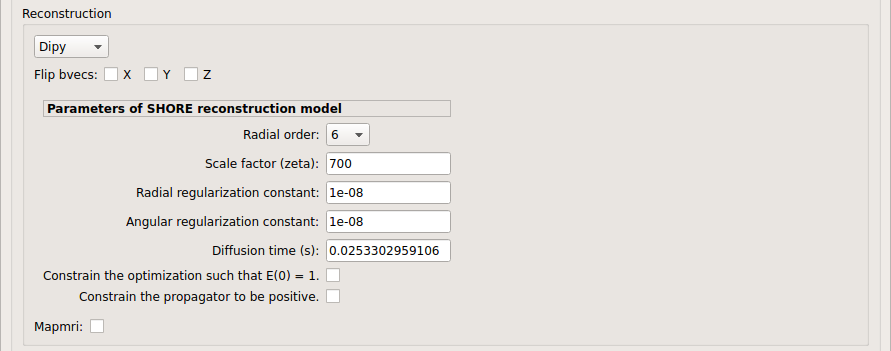
SHORE performed only on DSI data
Tensor:
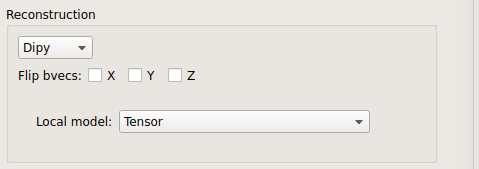
Tensor performed only on DTI data
CSD:
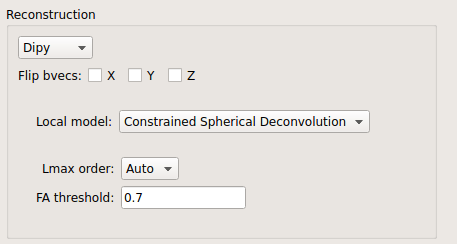
CSD performed on DTI and multi-shell data
MAP_MRI:
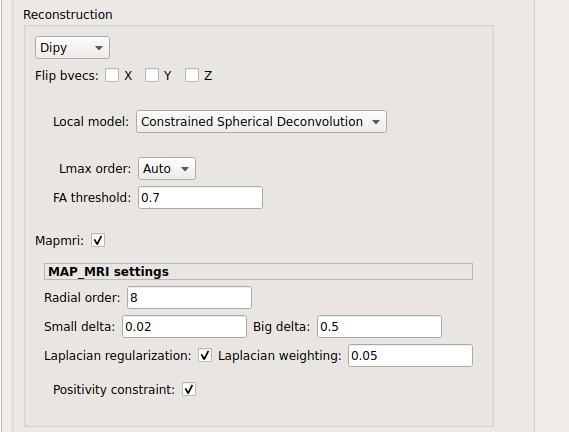
MAP-MRI performed only on multi-shell data
MRtrix: perform CSD reconstruction.
CSD:
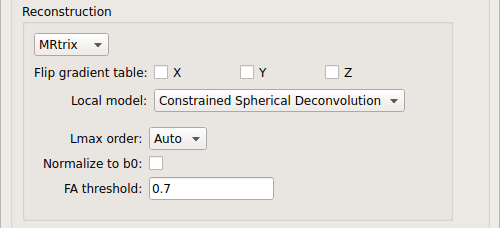
CSD performed on DTI and multi-shell data
Tractography tool
Dipy: perform deterministic and probabilistic fiber tracking as well as particle filtering tractography.
Deterministic tractography:
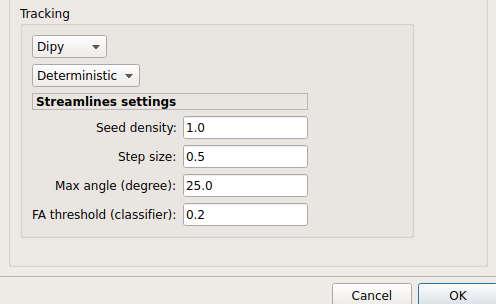
Deterministic tractography (SD_STREAM) performed on single tensor or CSD reconstruction
Probabilistic tractography:
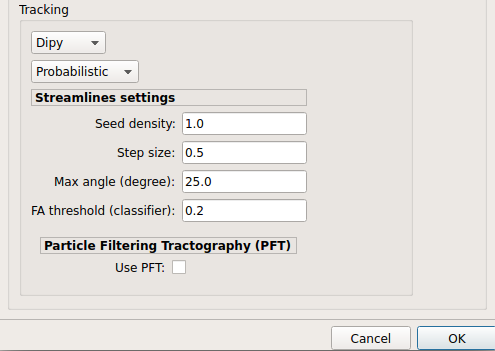
Probabilistic tractography (iFOD2) performed on SHORE or CSD reconstruction
Probabilistic particle filtering tractography (PFT):
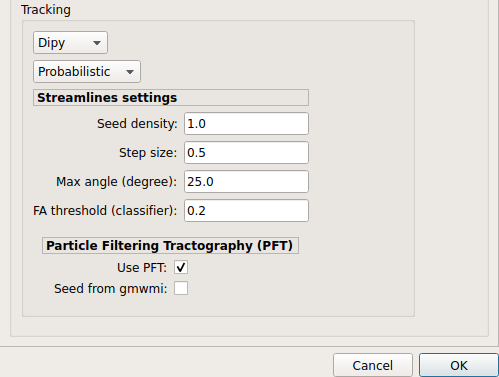
Probabilistic PFT tracking performed on SHORE or CSD reconstruction. Seeding from the gray matter / white matter interface is possible.
Note
We noticed a shift of the center of tractograms obtained by dipy. As a result, tractograms visualized in TrackVis are not commonly centered despite the fact that the tractogram and the ROIs are properly aligned.
MRtrix: perform deterministic and probabilistic fiber tracking as well as anatomically-constrained tractography. ROI dilation is required to map brain connections when the tracking only operates in the white matter.
Deterministic tractography:
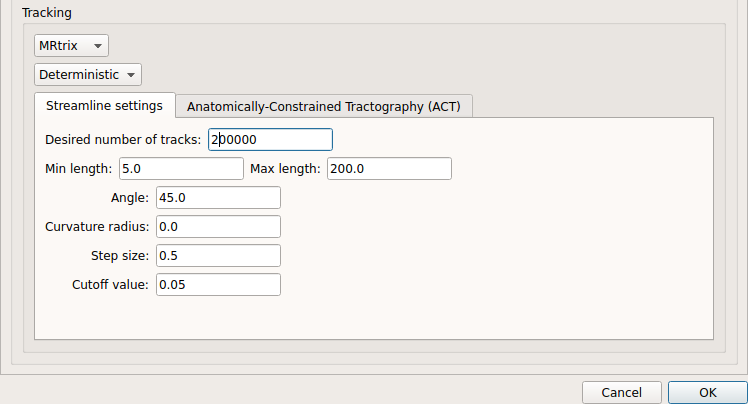
Deterministic tractography (SD_STREAM) performed on single tensor or CSD reconstruction
Deterministic anatomically-constrained tractography (ACT):
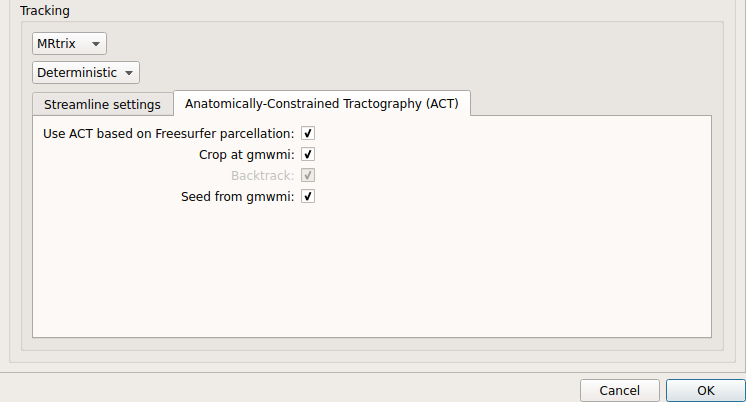
Deterministic ACT tracking performed on single tensor or CSD reconstruction. Seeding from the gray matter / white matter interface is possible. Backtrack option is not available in deterministic tracking.
Probabilistic tractography:
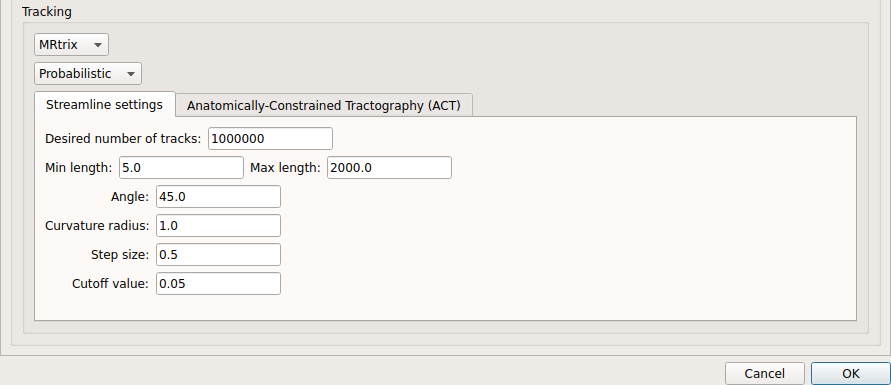
Probabilistic tractography (iFOD2) performed on SHORE or CSD reconstruction
Probabilistic anatomically-constrained tractography (ACT):
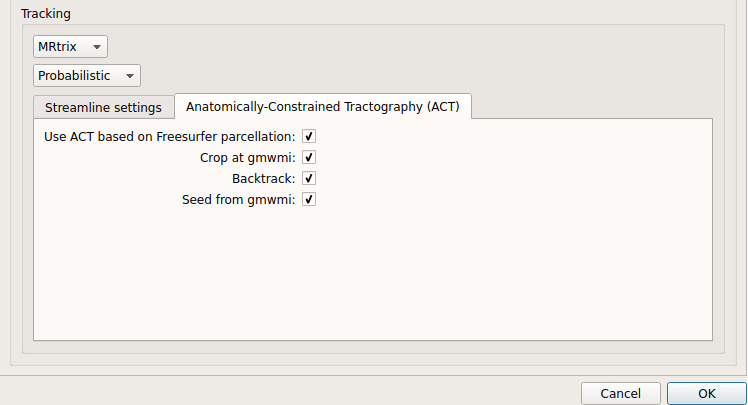
Probabilistic ACT tracking performed on SHORE or CSD reconstruction. Seeding from the gray matter / white matter interface is possible.
Connectome¶
Compute fiber length connectivity matrices. If DTI data is processed, FA additional map is computed. In case of DSI, additional maps include GFA and RTOP. In case of MAP-MRI, additional maps are RTPP, RTOP, …
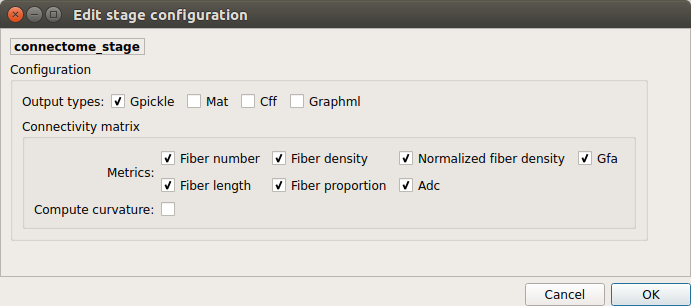
Output types
Select in which formats the connectivity matrices should be saved.
FMRI pipeline stages¶
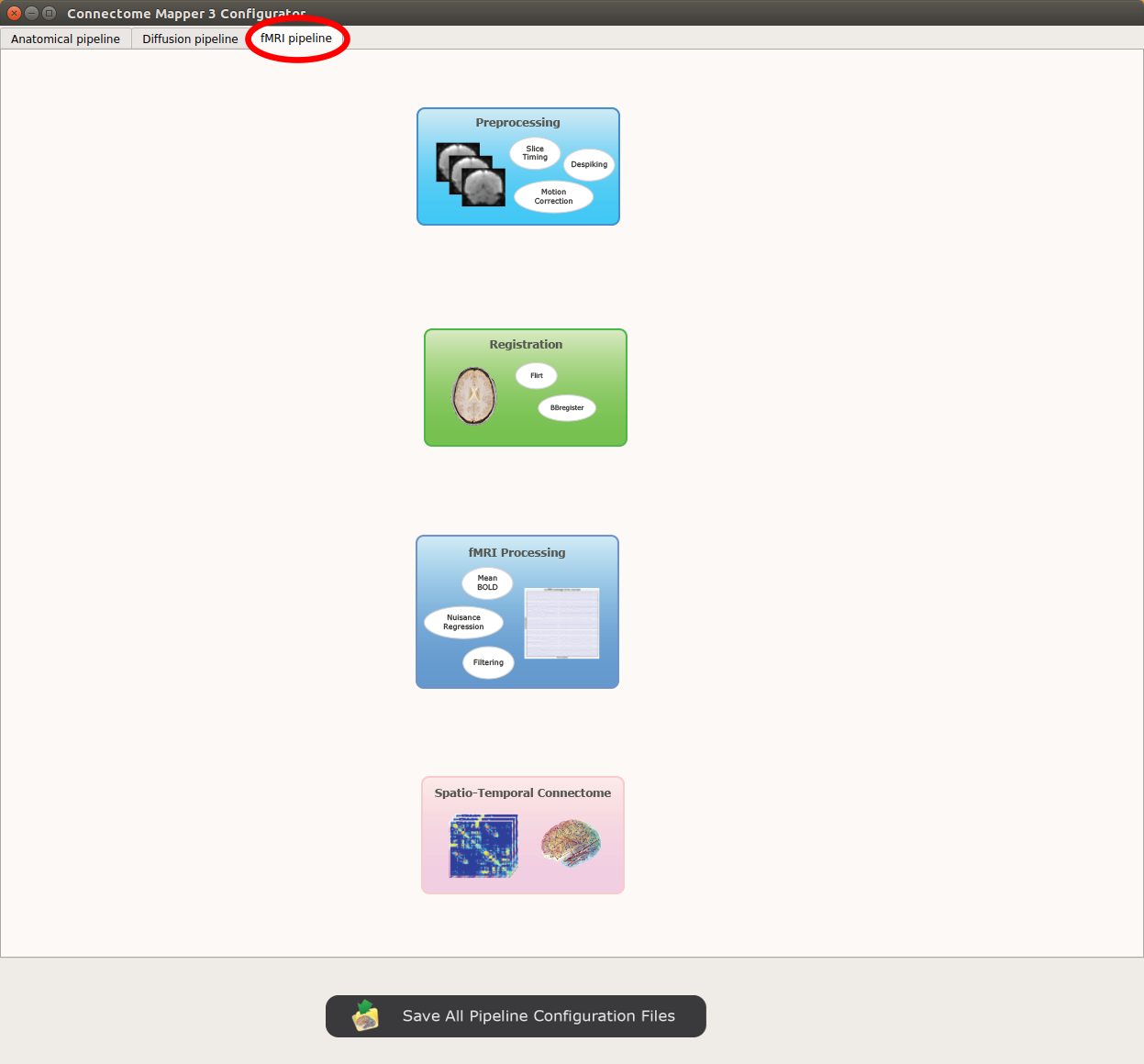
Panel for configuration of fMRI pipeline stages
Preprocessing¶
Preprocessing refers to processing steps prior to registration. It includes discarding volumes, despiking, slice timing correction and motion correction for fMRI (BOLD) data.
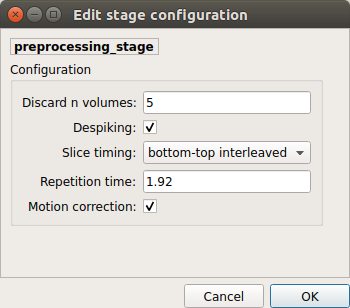
Discard n volummes
Discard n volumes from further analysis
Despiking
Perform despiking of the BOLD signal using AFNI.
Slice timing and Repetition time
Perform slice timing correction using FSL’s slicetimer.
Motion correction
Align BOLD volumes to the mean BOLD volume using FSL’s MCFLIRT.
Registration¶
Registration mode
FSL (Linear):
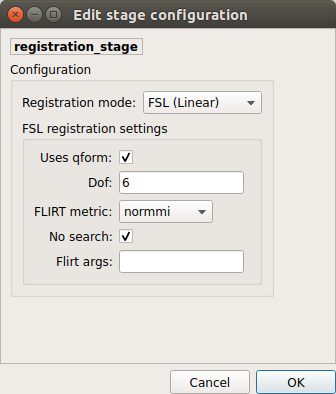
Perform linear registration from T1 to mean BOLD using FSL’s flirt.
BBregister (FS)
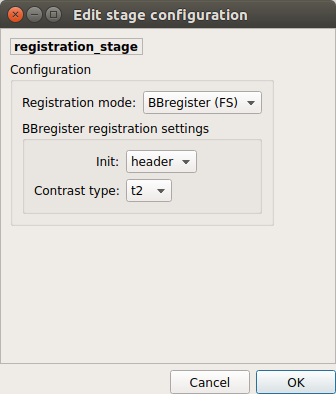
Perform linear registration using Freesurfer BBregister tool from T1 to mean BOLD via T2.
Warning
development in progress
fMRI processing¶
Performs detrending, nuisance regression, bandpass filteringdiffusion reconstruction and local deterministic or probabilistic tractography based on several tools. ROI dilation is required to map brain connections when the tracking only operates in the white matter.
Detrending
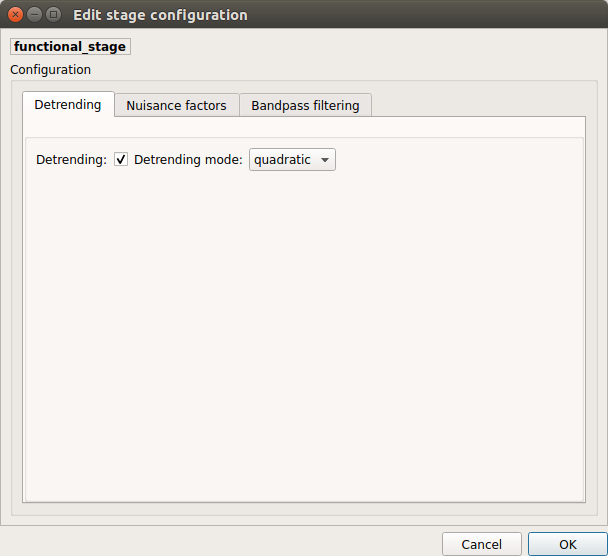
Detrending of BOLD signal using: 1. linear trend removal algorithm provided by the
scipylibrary 2. quadratic trend removal algorithm provided by theobspylibrary
Nuisance regression
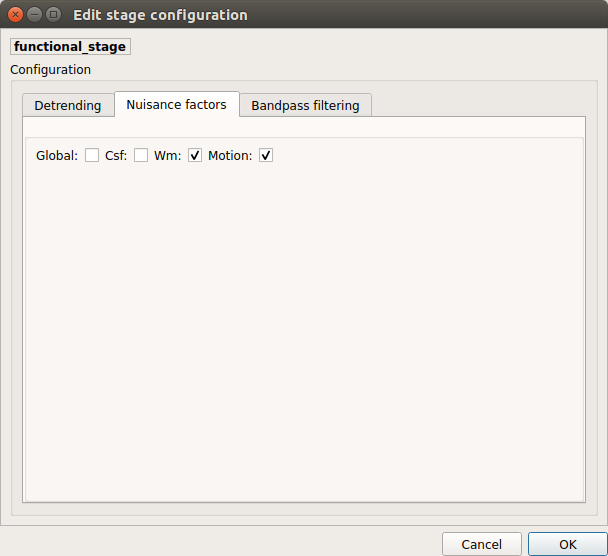
A number of options for removing nuisance signals is provided. They consist of: 1. Global signal regression 2. CSF regression 3. WM regression 4. Motion parameters regression
Bandpass filtering
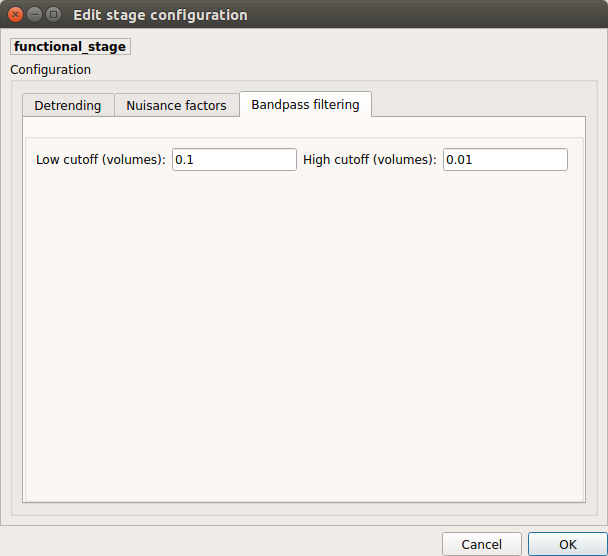
Perform bandpass filtering of the time-series using FSL’s slicetimer
Connectome¶
Computes ROI-averaged time-series and the correlation connectivity matrices.
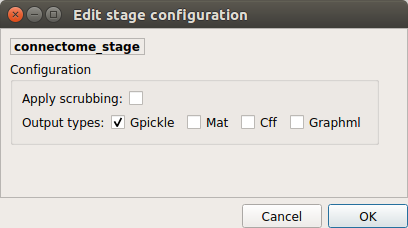
Output types
Select in which formats the connectivity matrices should be saved.
Save the configuration files¶
You can save the pipeline stage configuration files in two different way:
- You can save all configuration files at once by clicking on the
Save All Pipeline Configuration Files. This will save automatically the configuration file of the anatomical / diffusion / fMRI pipeline to<bids_dataset>/code/ref_anatomical_config.ini/<bids_dataset>/code/ref_diffusion_config.ini/<bids_dataset>/code/ref_fMRI_config.inirespectively.- You can save individually each of the pipeline configuration files and edit its filename in the File menu (File -> Save anatomical/diffusion/fMRI configuration file as…)
Nipype¶
The Connectome Mapper processing relies on nipype. For each stage, a processing folder is created in $Base_directory/derivatives/nipype/sub-<participant_label>/<pipeline_name>/<stage_name>.
All intermediate steps for the processing are saved in the corresponding stage folders.
Run the BIDS App¶
Start the Connectome Mapper BIDS App GUI¶
- From the main window, click on the middle button to start the Connectome Mapper BIDS App GUI.
- The window of the Connectome Mapper BIDS App GUI will appear, which will help you in setting up and launching the BIDS App run.
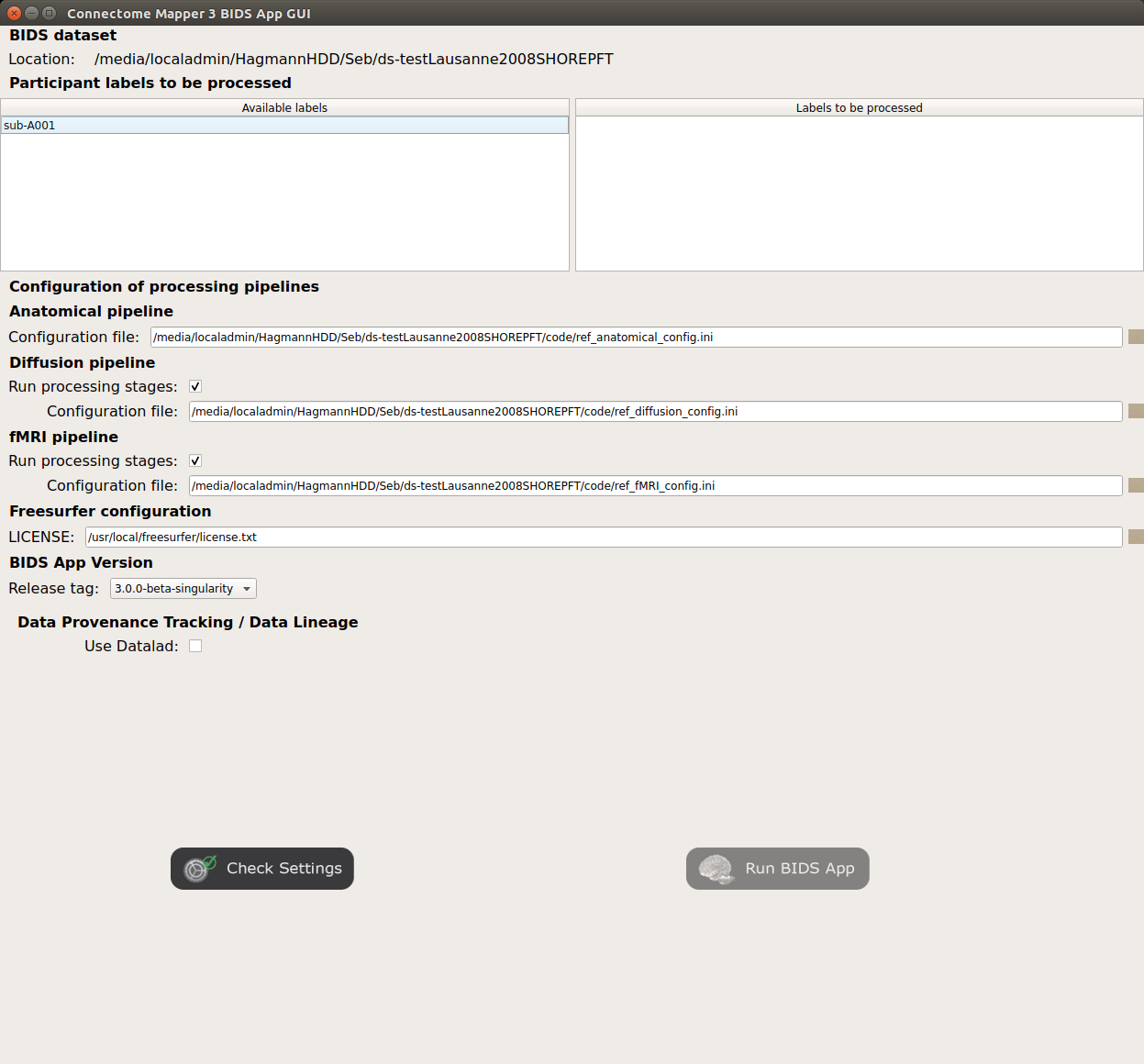
Window of the Connectome Mapper BIDS App GUI
Run configuration¶
Select the subject labels to be processed
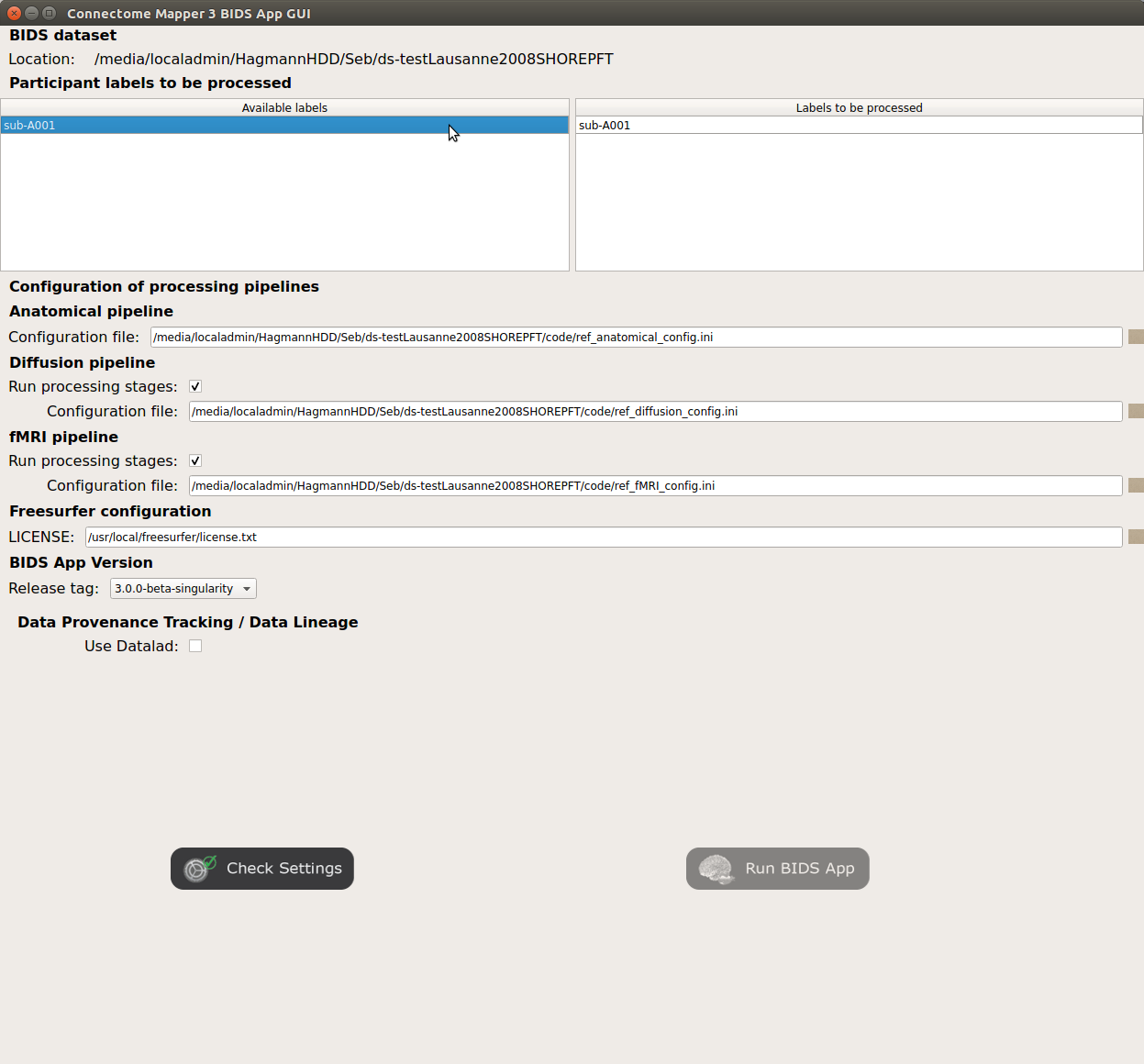
Check/Uncheck the pipelines to be performed
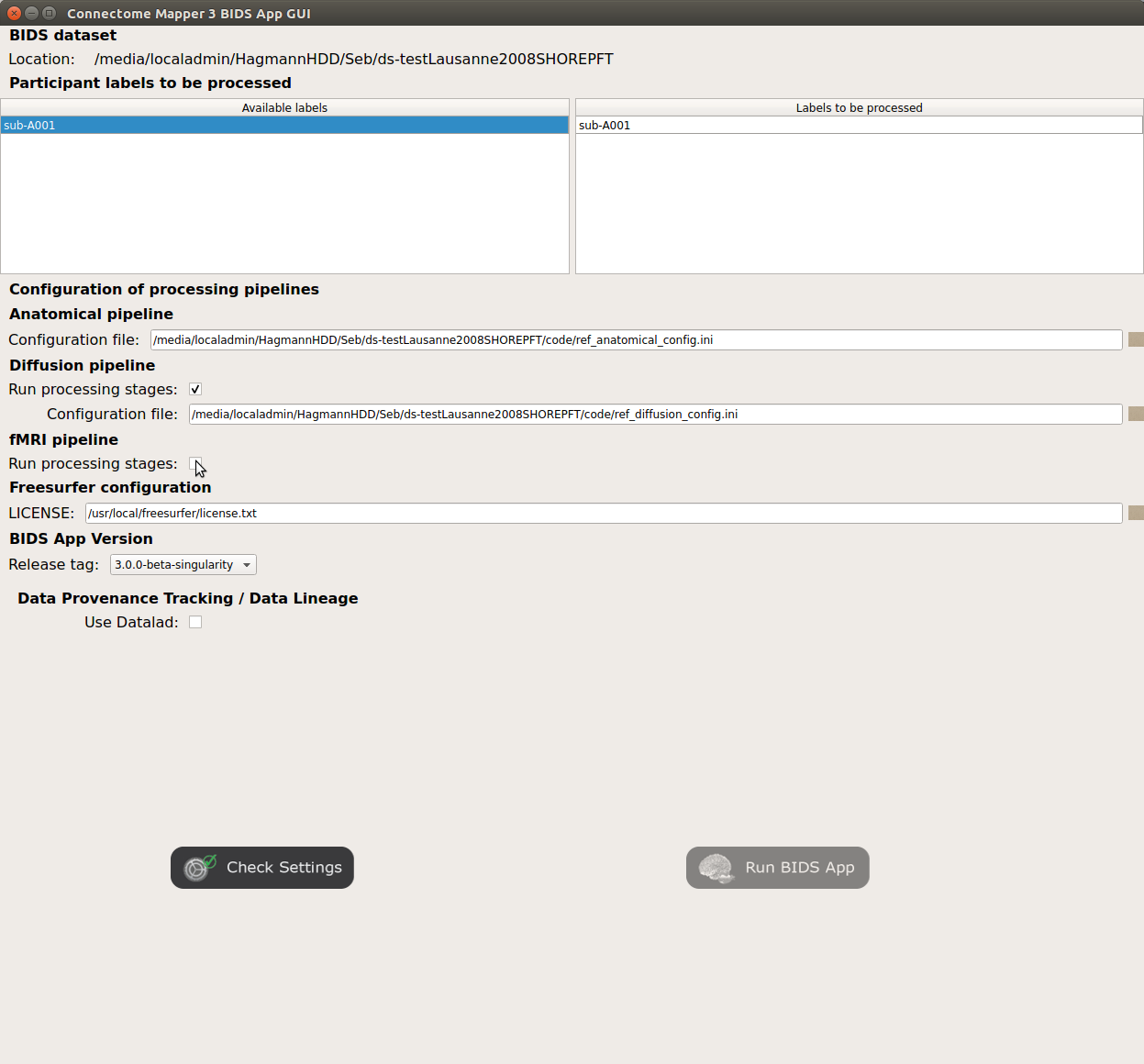
Specify your Freesurfer license
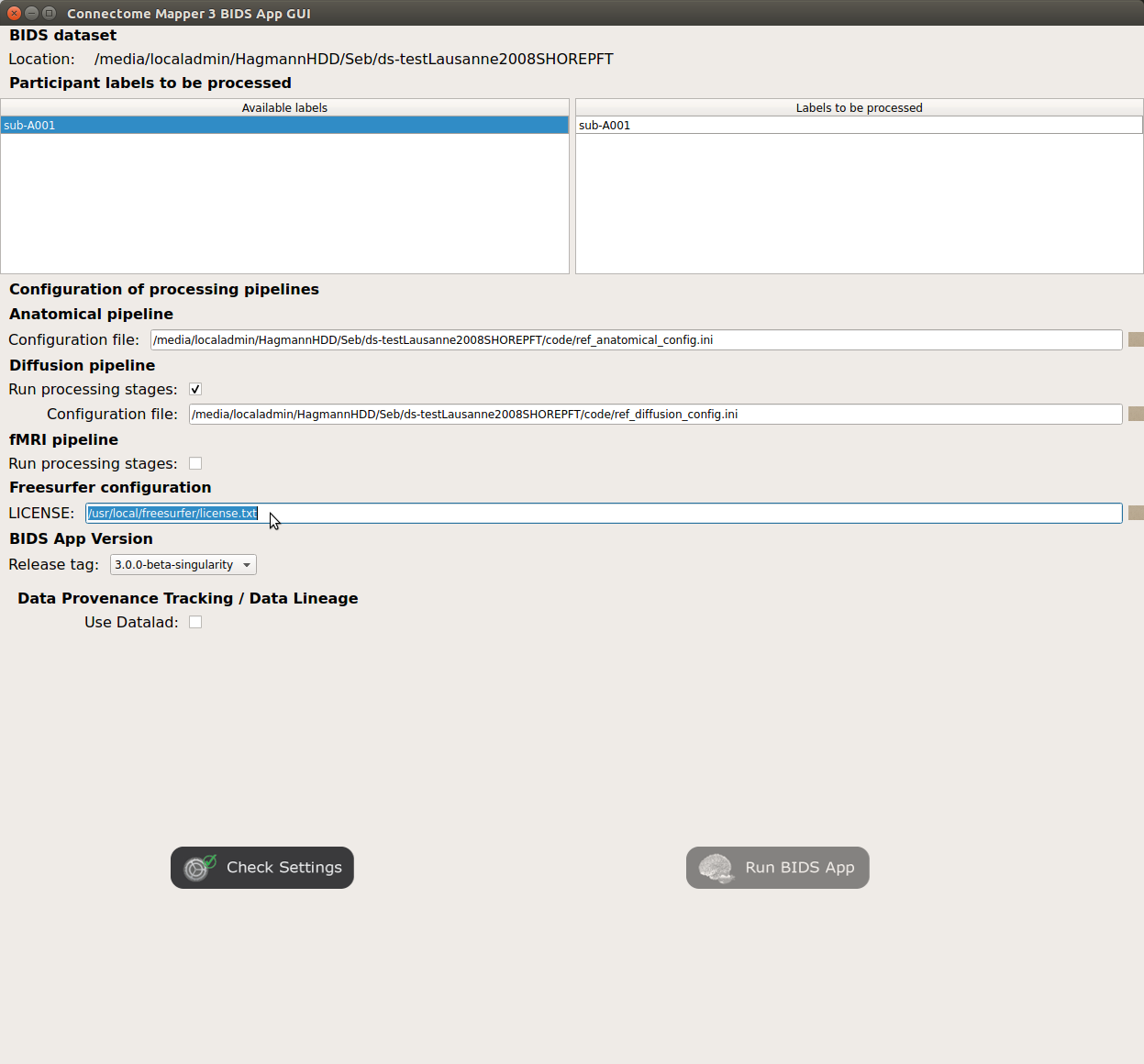
Note
Your freesurfer license will be copied to your dataset directory as
<bids_dataset>/code/license.txtwhich will be mounted inside the BIDS App container image.When the run is set up, you can click on the
Check settingsbutton.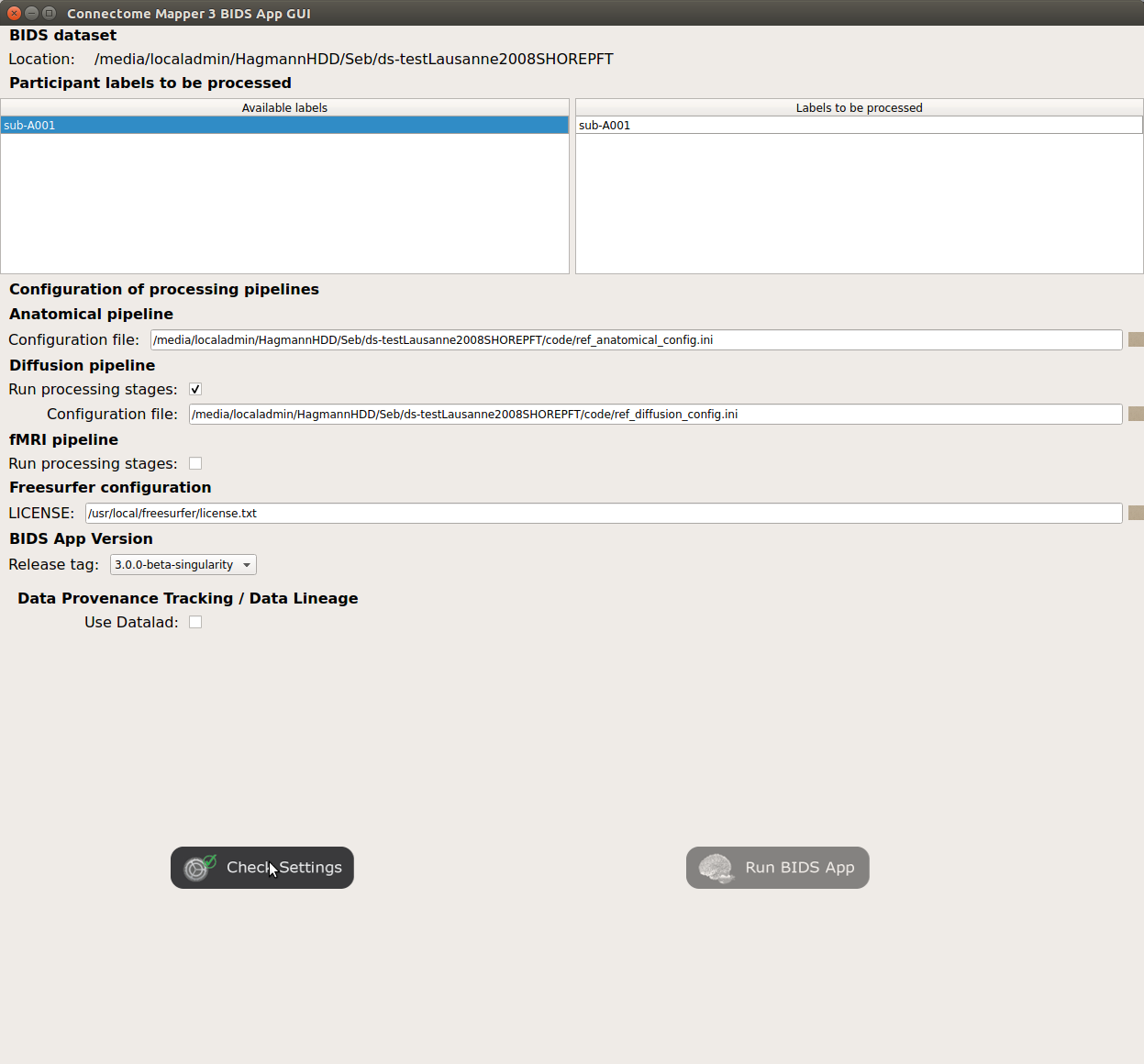
If the setup is complete and valid, this will enable the
Run BIDS Appbutton.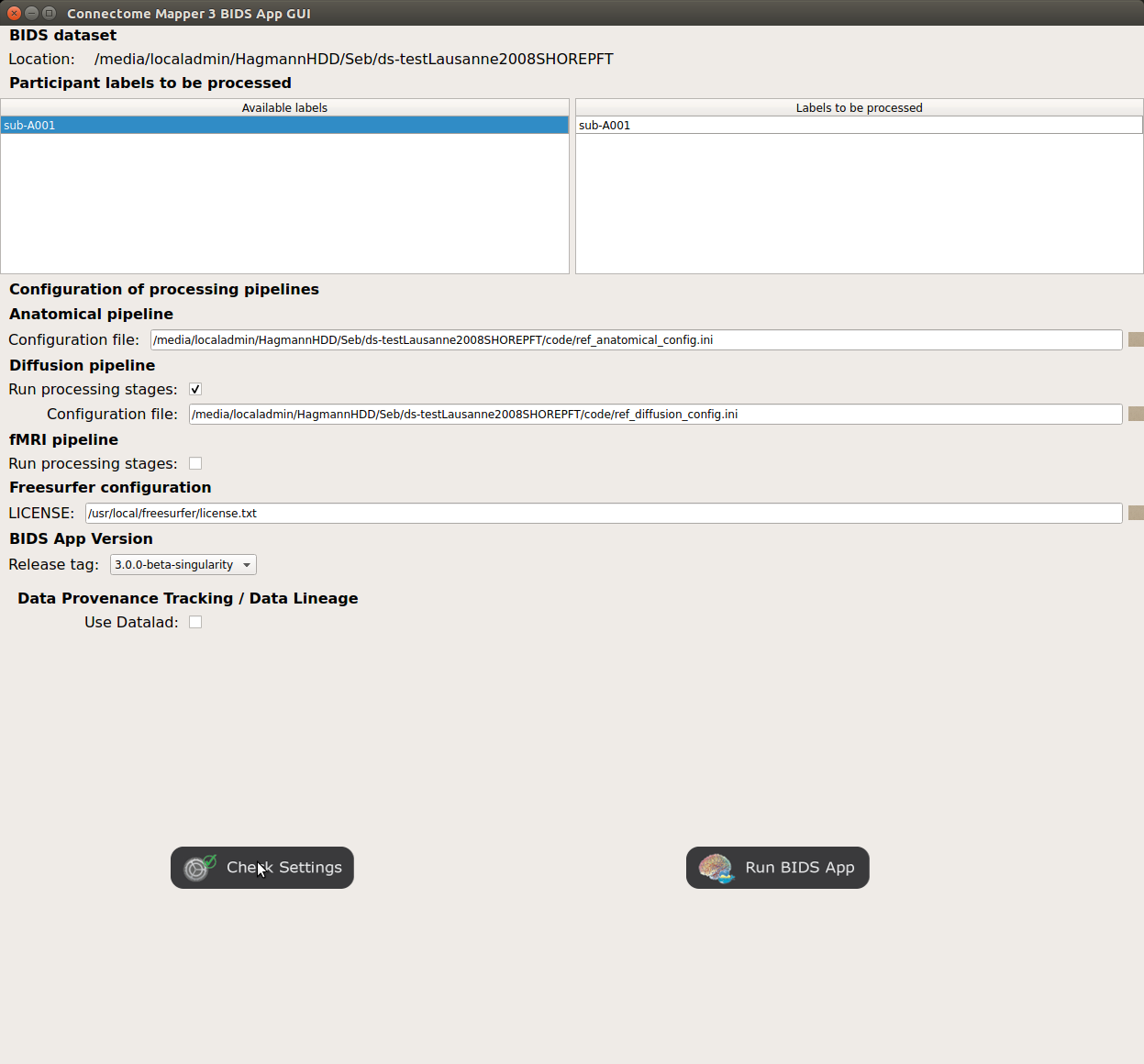
You are ready to launch the BIDS App run!
Launch the BIDS App run¶
Click on the
Run BIDS Appbutton to launch the BIDS App run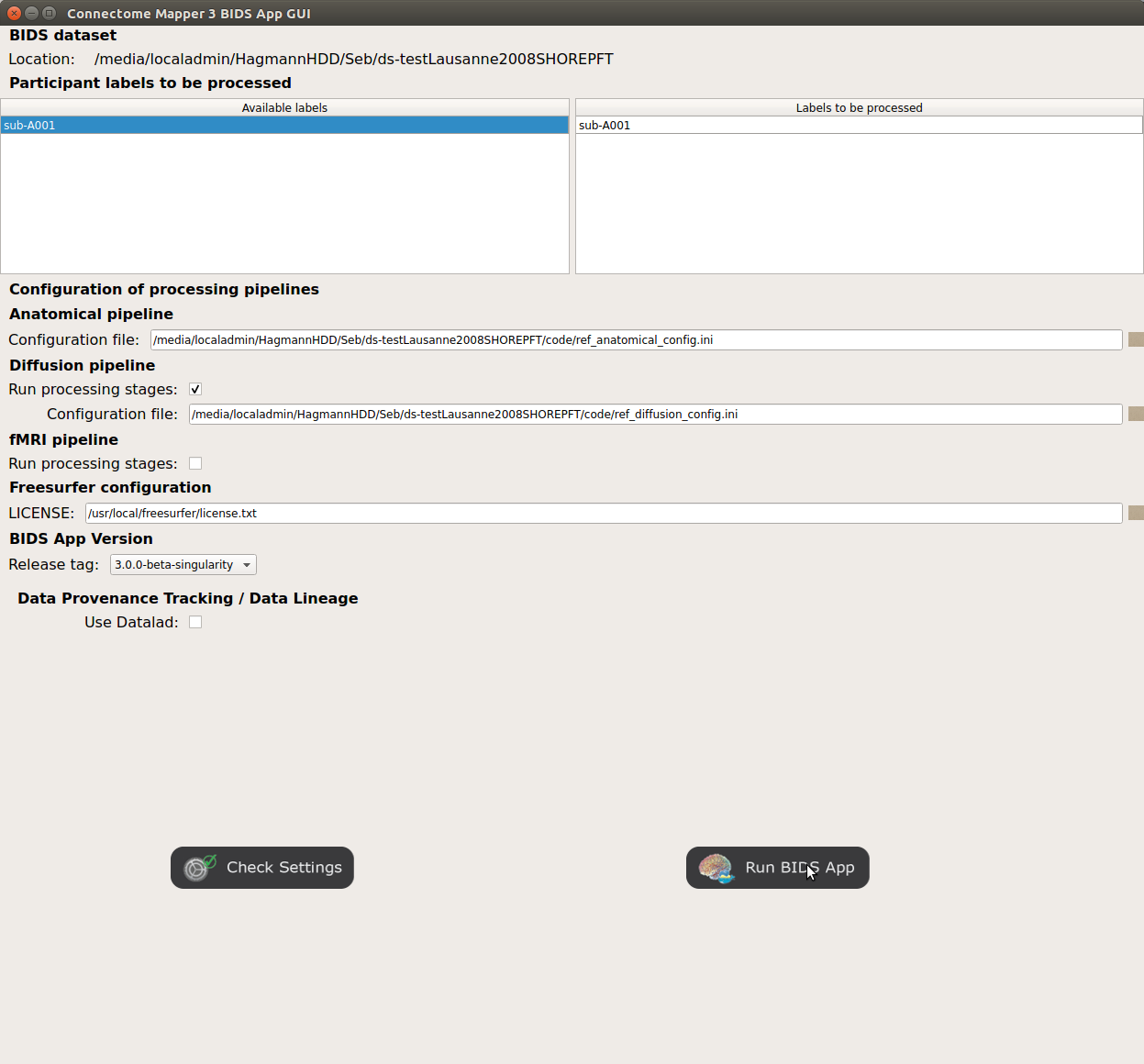
- You can see the complete
docker runcommand generated by the Connectome Mapper BIDS App GUI from the terminal output such as in this example Start BIDS App > Copy FreeSurfer license (BIDS App Manager) ... src : /usr/local/freesurfer/license.txt ... dst : /media/localadmin/HagmannHDD/Seb/ds-testLausanne2008SHOREPFT/code/license.txt > Datalad available: True *... Docker cmd 2 : ['docker', 'run', '-it', '--rm', '-v', '/media/localadmin/HagmannHDD/Seb/ds-testLausanne2008SHOREPFT:/tmp', '-u', '1000:1000', 'sebastientourbier/connectomemapper-bidsapp:3.0.0-beta-singularity', '/tmp', '/tmp/derivatives', 'participant', '--participant_label', 'A001', '--anat_pipeline_config', '/tmp/code/ref_anatomical_config.ini', '--dwi_pipeline_config', '/tmp/code/ref_diffusion_config.ini']* > BIDS dataset: /tmp > Subjects to analyze : ['A001'] > Copy FreeSurfer license (BIDS App) > Sessions to analyze : ['ses-20150203160809'] > Process subject sub-A001 session ses-20150203160809 WARNING: rewriting config file /tmp/derivatives/sub-A001_ses-20150203160809_anatomical_config.ini ... Anatomical config created : /tmp/derivatives/sub-A001_ses-20150203160809_anatomical_config.ini WARNING: rewriting config file /tmp/derivatives/sub-A001_ses-20150203160809_diffusion_config.ini ... Diffusion config created : /tmp/derivatives/sub-A001_ses-20150203160809_diffusion_config.ini ... Running pipelines : - Anatomical MRI (segmentation and parcellation) - Diffusion MRI (structural connectivity matrices) ... cmd : connectomemapper3 /tmp /tmp/derivatives sub-A001 ses-20150203160809 /tmp/derivatives/sub-A001_ses-20150203160809_anatomical_config.ini True /tmp/derivatives/sub-A001_ses-20150203160809_diffusion_config.ini True
Note
Also, this can be helpful in you wish to design your own batch scripts to call the BIDS App with the correct syntax.
- You can see the complete
Check progress¶
For each subject, the execution output of the pipelines are redirected to a log file, written as <bids_dataset/derivatives>/cmp/sub-<subject_label>_log.txt. Execution progress can be checked by the means of these log files.
Check stages outputs¶
Start the Inspector Window¶
- From the main window, click on the right button to start the Inspector Window.
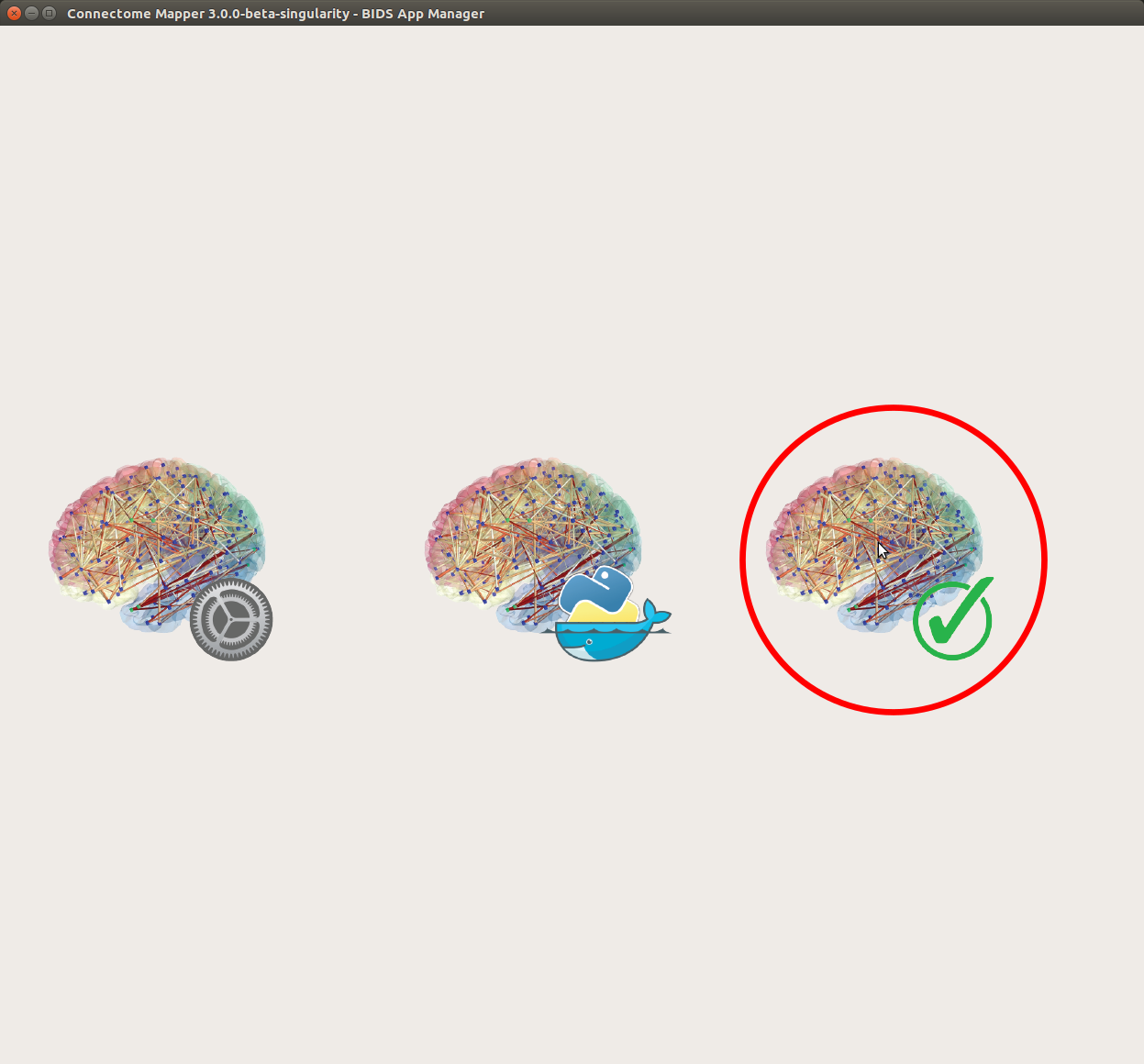
- The window of the Connectome Mapper BIDS App Inspector will appear, which will assist you in inspecting outputs of the different pipeline stages (each pipeline has a tab panel).
Anatomical pipeline stages¶
Click on the stage you wish to check the output(s):
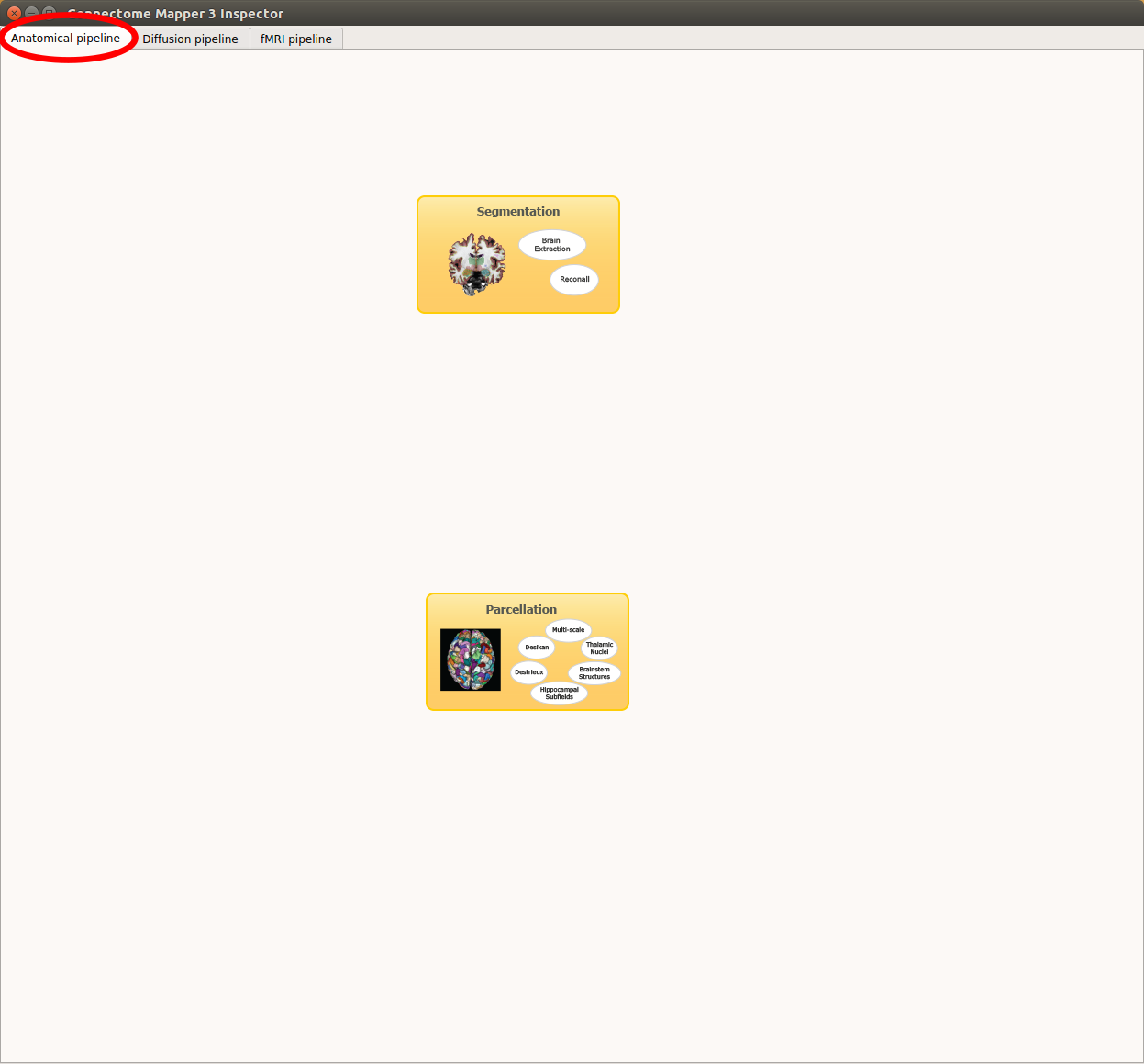
Panel for configuration of anatomical pipeline stages
Segmentation¶
Select the desired output from the list and click on
view:
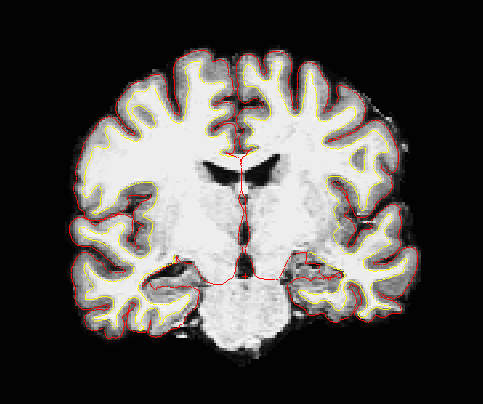
Segmentation results
Surfaces extracted using Freesurfer.
T1 segmented using Freesurfer.
Parcellation¶
Select the desired output from the list and click on
view:
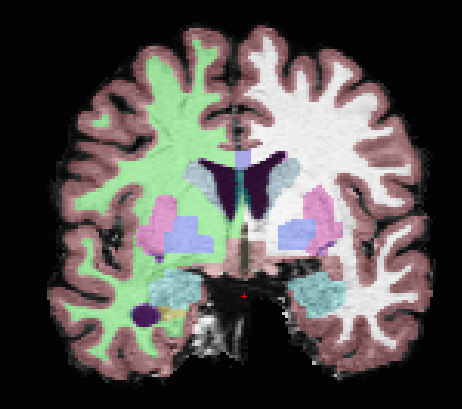
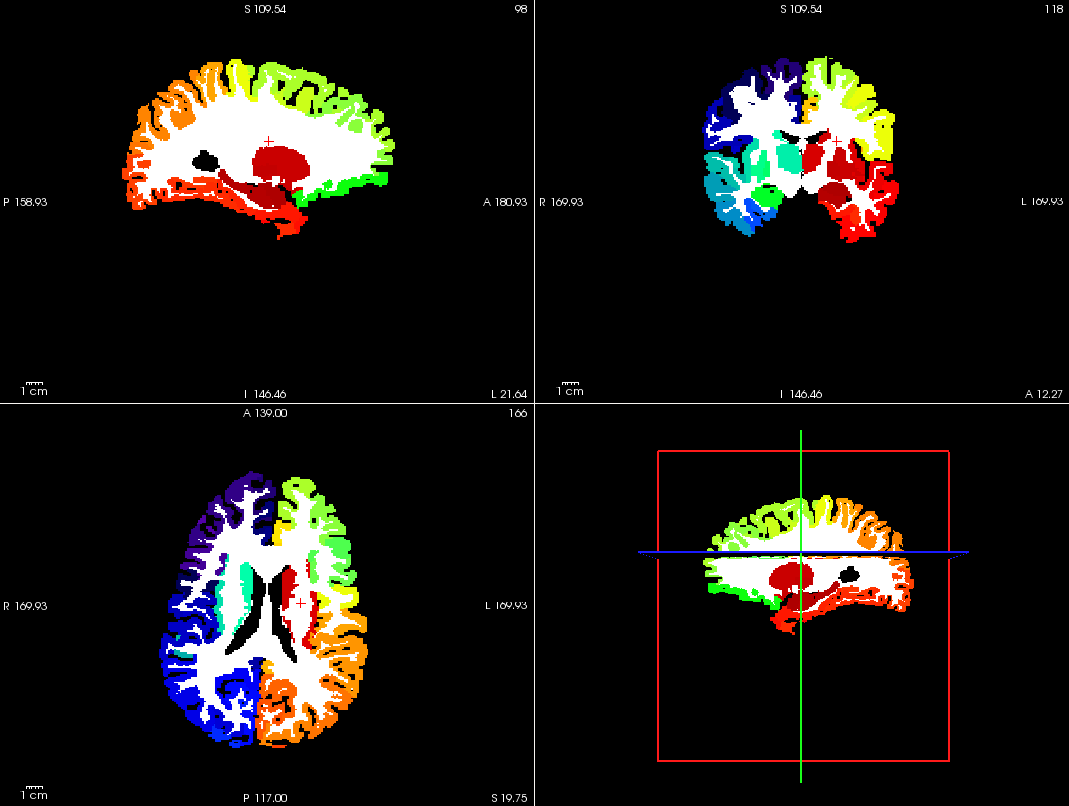
Parcellation results
Cortical and subcortical parcellation are shown with Freeview.
Diffusion pipeline stages¶
Click on the stage you wish to check the output(s):
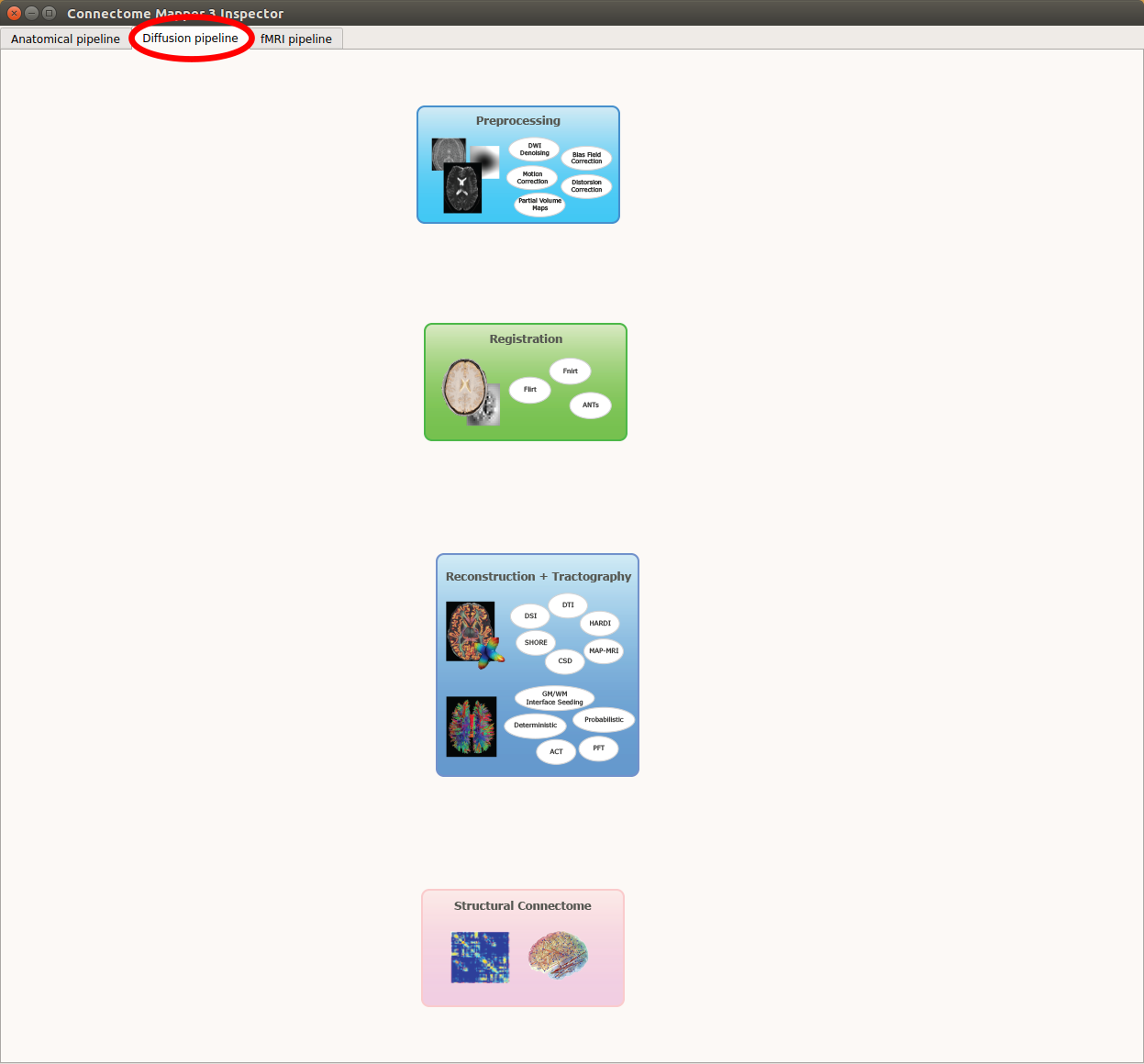
Panel for configuration of diffusion pipeline stages
Preprocessing¶
Select the desired output from the list and click on
view: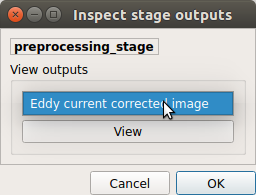
Registration¶
Select the desired output from the list and click on
view: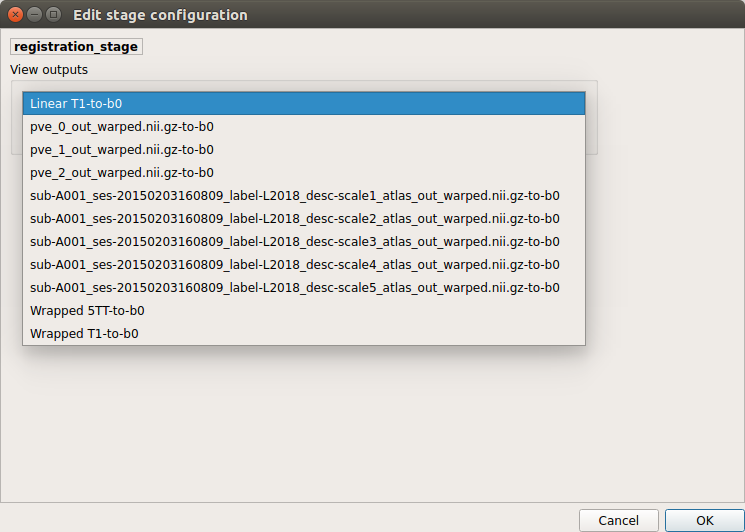
Registration results
Registration of T1 to Diffusion space (b0). T1 in copper overlayed to the b0 image.
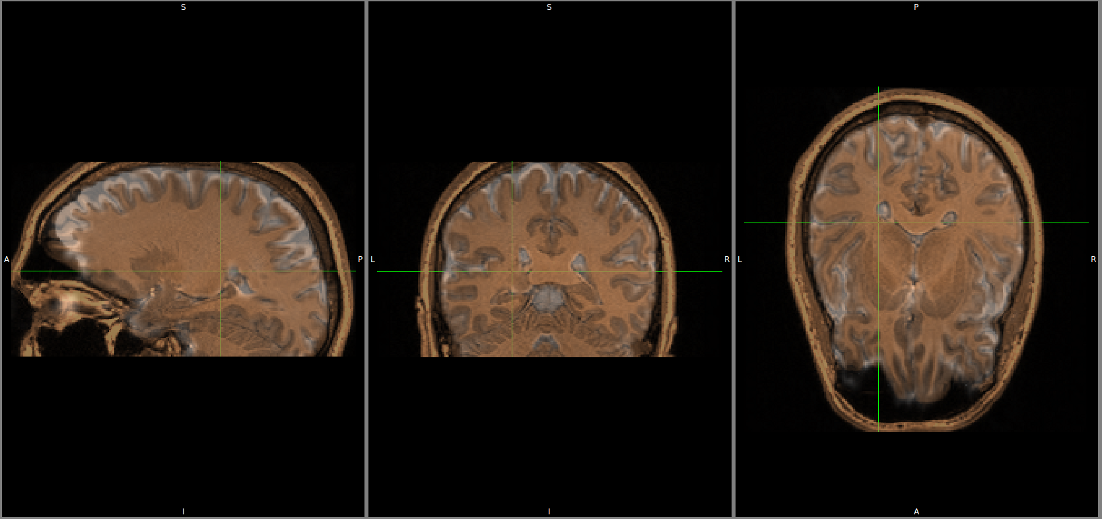
Diffusion reconstruction and tractography¶
Select the desired output from the list and click on
view: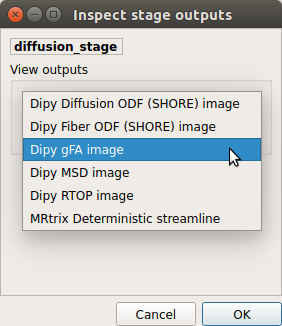
Tractography results
DSI Tractography results are displayed with TrackVis.
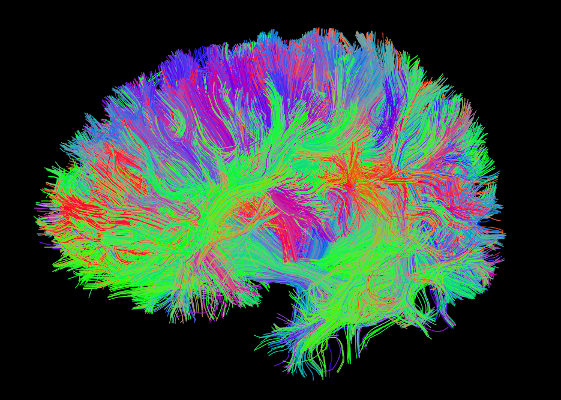
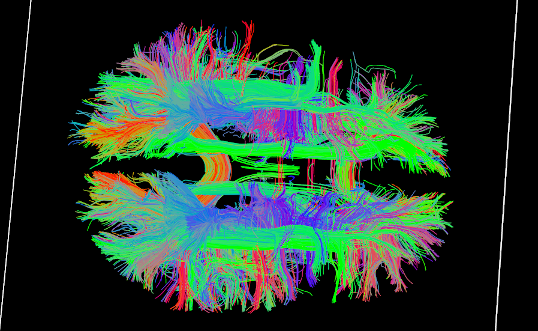
Connectome¶
Select the desired output from the list and click on
view: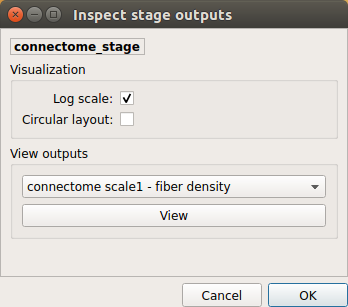
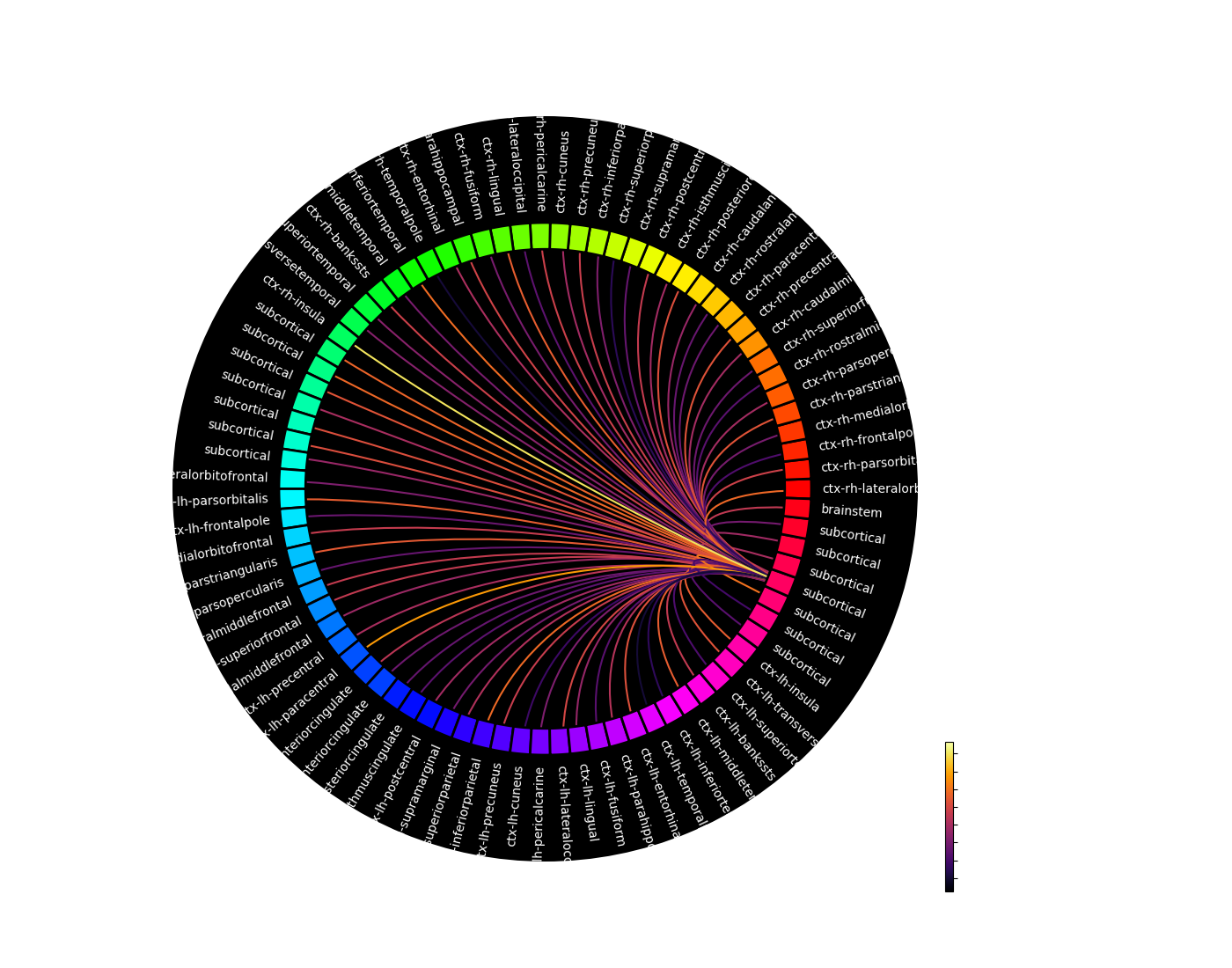
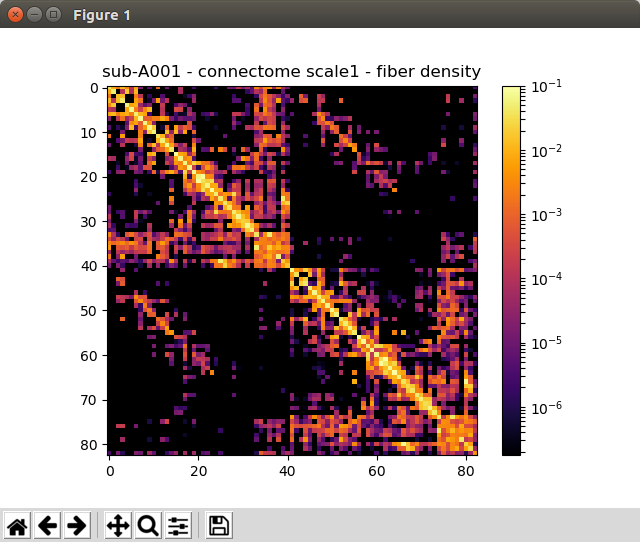
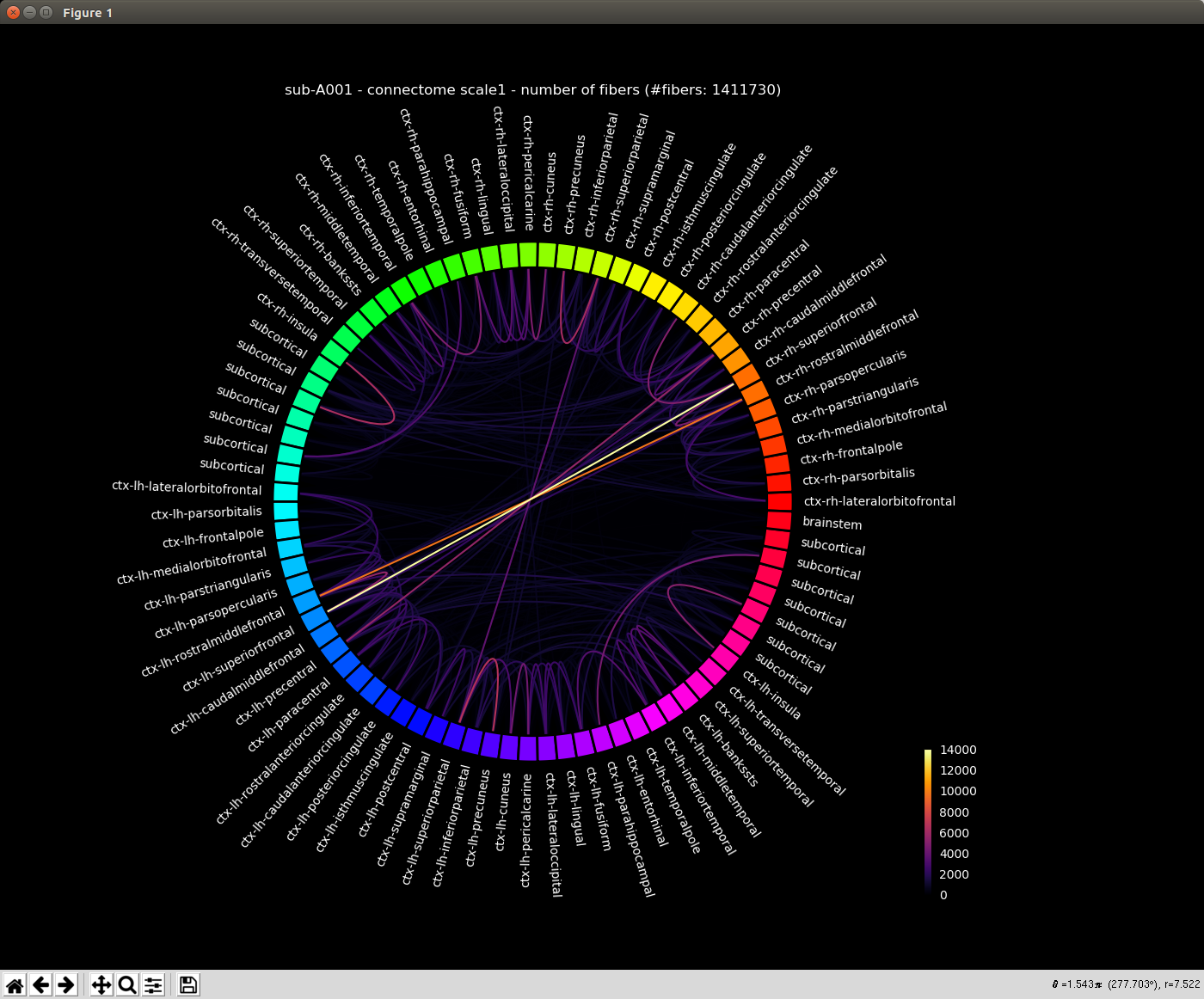
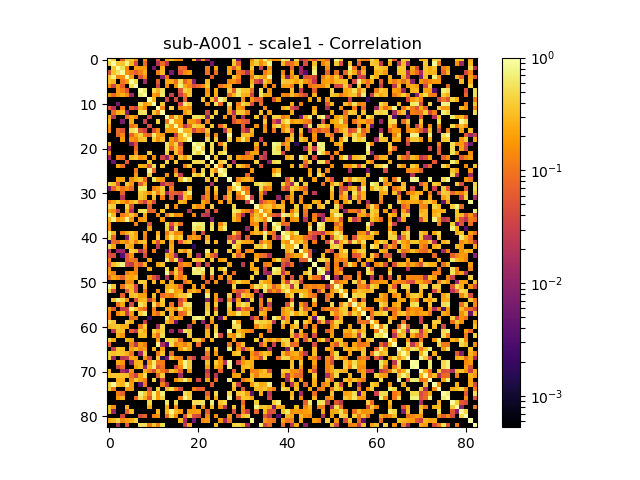
Generated connection matrix
Displayed using a:
- matrix layout with pyplot
- circular layout with pyplot and MNE
FMRI pipeline stages¶
Click on the stage you wish to check the output(s):
Panel for configuration of fMRI pipeline stages
Preprocessing¶
Select the desired output from the list and click on
view:
Registration¶
Select the desired output from the list and click on
view:
fMRI processing¶
Select the desired output from the list and click on
view:
ROI averaged time-series
Connectome¶
Select the desired output from the list and click on
view:
Generated connection matrix
Displayed using a:
- matrix layout with pyplot
- circular layout with pyplot and MNE