Connectome Mapper 3
Latest released version: v3.0.0
This neuroimaging processing pipeline software is developed by the Connectomics Lab at the University Hospital of Lausanne (CHUV) for use within the SNF Sinergia Project 170873, as well as for open-source software distribution. Source code is hosted on GitHub.





Warning
THIS SOFTWARE IS FOR RESEARCH PURPOSES ONLY AND SHALL NOT BE USED FOR ANY CLINICAL USE. THIS SOFTWARE HAS NOT BEEN REVIEWED OR APPROVED BY THE FOOD AND DRUG ADMINISTRATION OR EQUIVALENT AUTHORITY, AND IS FOR NON-CLINICAL, IRB-APPROVED RESEARCH USE ONLY. IN NO EVENT SHALL DATA OR IMAGES GENERATED THROUGH THE USE OF THE SOFTWARE BE USED IN THE PROVISION OF PATIENT CARE.
About
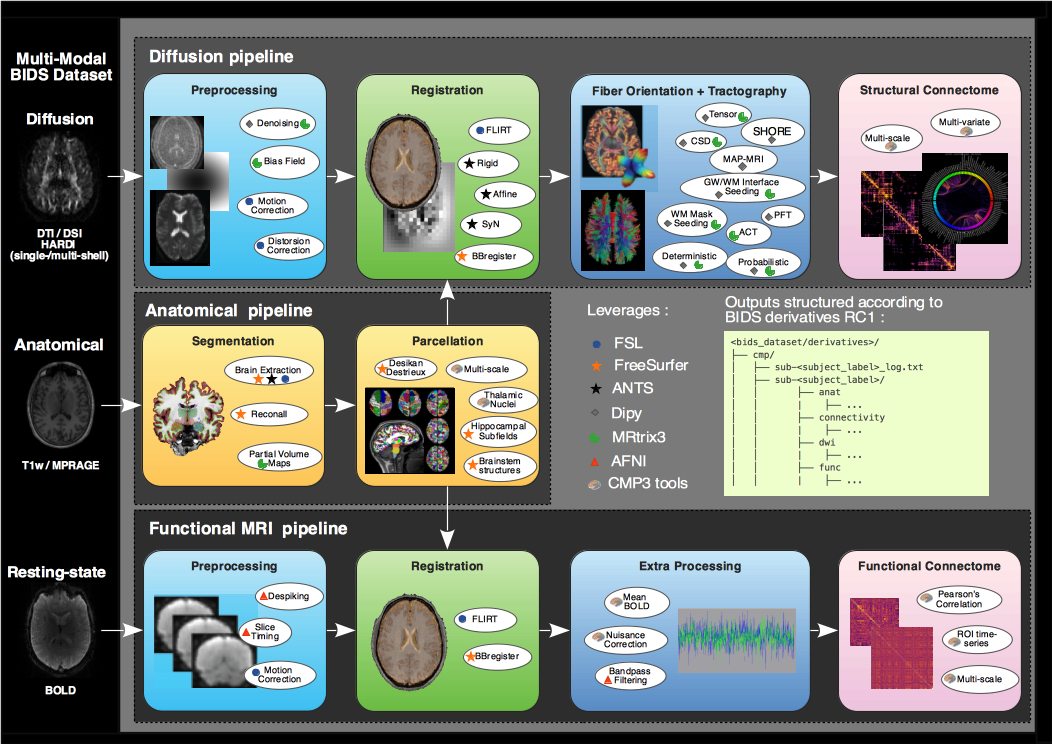
Connectome Mapper 3 is an open-source Python3 image processing pipeline software,
with a Graphical User Interface (GUI), that implements full anatomical, diffusion and
resting-state MRI processing pipelines, from raw Diffusion / T1 / T2 / BOLD data
to multi-resolution connection matrices based on a new version of the Lausanne
parcellation atlas, aka Lausanne2018.
Connectome Mapper 3 pipelines use a combination of tools from
well-known software packages, including FSL, FreeSurfer, ANTs,
MRtrix3, Dipy and AFNI, empowered by the Nipype dataflow library.
These pipelines are designed to provide the best software implementation
for each state of processing at the time of conception, and can be
easily updated as newer and better neuroimaging software become available.
Portability, reproducibility and replicatibility are achieved through the distribution of a BIDSApp, a software container image which (1) takes datasets organized following the Brain Imaging Data Structure (BIDS) standard, and which (2) provides a frozen computing environment where versions of all external softwares and libraries are fixed. Accessibility has been improved to a greater extend by providing an interactive GUI which supports the user in a the steps involved in the configuration and execution of the containerized pipelines.
This tool allows you to easily do the following:
Handle T1 / Diffusion / resting-state MRI data, organized following the Brain Imaging Data Structure (BIDS) standard, from raw to multi-resolution connection matrices.
Implement tools from different software packages.
Achieve optimal data processing quality by using the best tools available
Automate and parallelize processing steps with Nipype, which provides a significant speed-up from typical linear, manual processing.
Easily configure the pipelines and control the execution and outputs of the processing with a GUI.
License information
This software is distributed under the open-source license Modified BSD. See license for more details.
All trademarks referenced herein are property of their respective holders.
Aknowledgment
If your are using the Connectome Mapper 3 in your work, please acknowledge this software. See Citing for more details.
Help/Questions
If you run into any problems or have any questions, you can post to the CMTK-users group. Code bugs can be reported by creating a “New Issue” on the source code repository.
Eager to contribute?
Connectome Mapper 3 is open-source and all kind of contributions (bug reporting, documentation, code,…) are welcome! See Contributing to Connectome Mapper for more details.
Contents
Getting started
User Documentation
API Documentation
Examples & Tutorials
Funding
Work supported by the SNF Sinergia Grant 170873 (http://p3.snf.ch/Project-170873).