Outputs of Connectome Mapper 3
Processed, or derivative, data are outputed to <bids_dataset/derivatives>/.
BIDS derivatives entities
Entity |
Description |
|---|---|
|
Distinguish different subjects |
|
Distinguish different acquisition sessions |
|
Distinguish different experiment tasks |
|
Describe the type of brain tissue segmented (for _probseg/dseg) |
|
Distinguish data derived from different types of parcellation atlases |
|
Distinguish data derived from the different scales of Lausanne2008 and Lausanne2018 parcellation atlases |
|
Distinguish anatomical MRI derivatives in the target diffusion MRI space |
|
Distinguish different diffusion signal models (DTI, CSD, SHORE, MAPMRI) |
See Original BIDS Entities Appendix for more description.
Note
Connectome Mapper 3 introduced a new BIDS entity atlas-<atlas_label>
(where <atlas_label>: Desikan/ L2018), that is used
in combination with the res-<atlas_scale> (where <atlas_scale>:
scale1 / scale2 / scale3 / scale4 / scale5) entity to
distinguish data derived from different parcellation atlases and
different scales.
Main Connectome Mapper Derivatives
Main outputs produced by Connectome Mapper 3 are written to
cmp/sub-<subject_label>/. In this folder, a configuration file
generated for each modality pipeline (i.e. anatomical/diffusion/fMRI/EEG)
and used for processing each participant is saved as
sub-<subject_label>_anatomical/diffusion/fMRI/EEG_config.json.
It summarizes pipeline workflow options and parameters used for processing.
An execution log of the full workflow is saved as sub-<subject_label>_log.txt`.
Anatomical derivatives
Anatomical derivatives in the individual
T1wspace are placed in each subject’sanat/subfolder, including:The original T1w image:
anat/sub-<subject_label>_desc-head_T1w.nii.gz
The masked T1w image with its corresponding brain mask:
anat/sub-<subject_label>_desc-brain_T1w.nii.gzanat/sub-<subject_label>_desc-brain_mask.nii.gz
The segmentations of the white matter (WM), gray matter (GM), and Cortical Spinal Fluid (CSF) tissues:
anat/sub-<subject_label>_label-WM_dseg.nii.gzanat/sub-<subject_label>_label-GM_dseg.nii.gzanat/sub-<subject_label>_label-CSF_dseg.nii.gz
The five different brain parcellations:
anat/sub-<subject_label>_atlas-<atlas_label>[_res-<scale_label>]_dseg.nii.gzwhere:
<atlas_label>:Desikan/L2018is the parcellation scheme used<scale_label>:scale1,scale2,scale3,scale4,scale5corresponds to the parcellation scale if applicable
with two
tsvside-car files that follow the BIDS derivatives, one describing the parcel label/index mapping (_dseg.tsv), one reporting volumetry of the different parcels (_stats.tsv), and two files used internally by CMP3, one describing the parcel labels ingraphmlformat (dseg.graphml), one providing the color lookup table of the parcel labels in Freesurfer format which can used directly infreeview(_FreeSurferColorLUT.txt):anat/sub-<subject_label>_atlas-<atlas_label>[_res-<scale_label>]_dseg.tsvanat/sub-<subject_label>_atlas-<atlas_label>[_res-<scale_label>]_stats.tsvanat/sub-<subject_label>_atlas-<atlas_label>[_res-<scale_label>]_dseg.graphmlanat/sub-<subject_label>_atlas-<atlas_label>[_res-<scale_label>]_FreeSurferColorLUT.txt
Anatomical derivatives in the
DWIspace produced by the diffusion pipeline are placed in each subject’sanat/subfolder, including:The unmasked T1w image:
anat/sub-<subject_label>_space-DWI_desc-head_T1w.nii.gz
The masked T1w image with its corresponding brain mask:
anat/sub-<subject_label>_space-DWI_desc-brain_T1w.nii.gzanat/sub-<subject_label>_space-DWI_desc-brain_mask.nii.gz
The segmentation of WM tissue used for tractography seeding:
anat/sub-<subject_label>_space-DWI_label-WM_dseg.nii.gz
The five different brain parcellation are saved as:
anat/sub-<subject_label>_space-DWI_atlas-<atlas_label>[_res-<scale_label>]_dseg.nii.gzwhere:
<atlas_label>:Desikan/L2018is the parcellation scheme used<scale_label>:scale1,scale2,scale3,scale4,scale5corresponds to the parcellation scale if applicable
The 5TT image used for Anatomically Constrained Tractorgaphy (ACT):
anat/sub-<subject_label>_space-DWI_label-5TT_probseg.nii.gz
The patial volume maps for white matter (WM), gray matter (GM), and Cortical Spinal Fluid (CSF) used for Particale Filtering Tractography (PFT), generated from 5TT image:
anat/sub-<subject_label>_space-DWI_label-WM_probseg.nii.gzanat/sub-<subject_label_space-DWI>_label-GM_probseg.nii.gzanat/sub-<subject_label>_space-DWI_label-CSF_probseg.nii.gz
The GM/WM interface used for ACT and PFT seeding:
anat/sub-<subject_label>_space-DWI_label-GMWMI_probseg.nii.gz
Diffusion derivatives
Diffusion derivatives in the individual DWI space are placed in
each subject’s dwi/ subfolder, including:
The final preprocessed DWI image used to fit the diffusion model for tensor or fiber orientation distribution estimation:
dwi/sub-<subject_label>_desc-preproc_dwi.nii.gz
The brain mask used to mask the DWI image:
dwi/sub-<subject_label>_desc-brain_mask_resampled.nii.gz
The diffusion tensor (DTI) fit (if used for tractography):
dwi/sub-<subject_label>]_desc-WLS_model-DTI_diffmodel.nii.gzwith derived Fractional Anisotropic (FA) and Mean Diffusivity (MD) maps:
dwi/sub-<subject_label>]_model-DTI_FA.nii.gzdwi/sub-<subject_label>]_model-DTI_MD.nii.gz
The Fiber Orientation Distribution (FOD) image from Constrained Spherical Deconvolution (CSD) fit (if performed):
dwi/sub-<subject_label>]_model-CSD_diffmodel.nii.gz
The MAP-MRI fit for DSI and multi-shell DWI data (if performed):
dwi/sub-<subject_label>]_model-MAPMRI_diffmodel.nii.gz
with derived Generalized Fractional Anisotropic (GFA), Mean Squared Displacement (MSD), Return-to-Origin Probability (RTOP) and Return-to-Plane Probability (RTPP) maps:
dwi/sub-<subject_label>]_model-MAPMRI_GFA.nii.gzdwi/sub-<subject_label>]_model-MAPMRI_MSD.nii.gzdwi/sub-<subject_label>]_model-MAPMRI_RTOP.nii.gzdwi/sub-<subject_label>]_model-MAPMRI_RTPP.nii.gz
The SHORE fit for DSI data:
dwi/sub-<subject_label>]_model-SHORE_diffmodel.nii.gz
with derived Generalized Fractional Anisotropic (GFA), Mean Squared Displacement (MSD), Return-to-Origin Probability (RTOP) maps:
dwi/sub-<subject_label>]_model-SHORE_GFA.nii.gzdwi/sub-<subject_label>]_model-SHORE_MSD.nii.gzdwi/sub-<subject_label>]_model-SHORE_RTOP.nii.gz
The tractogram:
dwi/sub-<subject_label>_model-<model_label>_desc-<label>_tractogram.trkwhere:
<model_label>is the diffusion model used to drive tractography (DTI, CSD, SHORE)<label>is the type of tractography algorithm employed (DET for deterministic, PROB for probabilistic)
The structural connectivity (SC) graphs:
dwi/sub-<subject_label>_atlas-<atlas_label>[_res-<scale_label>]_conndata-network_connectivity.<fmt>where:
<atlas_label>:Desikan/L2018is the parcellation scheme used<scale_label>:scale1,scale2,scale3,scale4,scale5corresponds to the parcellation scale if applicable<fmt>:mat/gpickle/tsv/graphmlis the format used to store the graph
Functional derivatives
Functional derivatives in the ‘meanBOLD’ (individual) space are placed in
each subject’s func/ subfolder including:
The original BOLD image:
func/sub-<subject_label>_task-rest_desc-cmp_bold.nii.gz
The mean BOLD image:
func/sub-<subject_label>_meanBOLD.nii.gz
The fully preprocessed band-pass filtered used to compute ROI time-series:
func/sub-<subject_label>_desc-bandpass_task-rest_bold.nii.gz
For scrubbing (if enabled):
The change of variance (DVARS):
func/sub-<subject_label>_desc-scrubbing_DVARS.npy
The frame displacement (FD):
func/sub-<subject_label>_desc-scrubbing_FD.npy
Motion-related time-series:
func/sub-<subject_label>_motion.tsv
The ROI time-series for each parcellation scale:
func/sub-<subject_label>_atlas-<atlas_label>[_res-<scale_label>]_timeseries.npyfunc/sub-<subject_label>_atlas-<atlas_label>[_res-<scale_label>]_timeseries.matwhere:
<atlas_label>:Desikan/L2018is the parcellation scheme used<scale_label>:scale1,scale2,scale3,scale4,scale5corresponds to the parcellation scale if applicable
The functional connectivity (FC) graphs:
func/sub-<subject_label>_atlas-<atlas_label>[_res-<scale_label>]_conndata-network_connectivity.<fmt>where:
<atlas_label>:Desikan/L2018is the parcellation scheme used<scale_label>:scale1,scale2,scale3,scale4,scale5corresponds to the parcellation scale if applicable<fmt>:mat/gpickle/tsv/graphmlis the format used to store the graph
EEG derivatives
EEG derivatives are placed in each subject’s eeg/ subfolder including:
The preprocessed EEG epochs data in
fifformat:eeg/sub-<subject_label>_task-<task_label>_epo.fif
The BEM surfaces in
fifformat:eeg/sub-<subject_label>_task-<task_label>_bem.fif
The source space in
fifformat:eeg/sub-<subject_label>_task-<task_label>_src.fif
The forward solution in
fifformat:eeg/sub-<subject_label>_task-<task_label>_fwd.fif
The inverse operator in
fifformat:eeg/sub-<subject_label>_task-<task_label>_inv.fif
The computed noise covariance in
fifformat:eeg/sub-<subject_label>_task-<task_label>_noisecov.fif
The transform of electrode positions that might be used for ESI in
fifformat:eeg/sub-<subject_label>_trans.fif
The ROI time-series for each parcellation atlas (and scale):
eeg/sub-<subject_label>_task-<task_label>_atlas-<atlas_label>[_res-<scale_label>]_timeseries.npyeeg/sub-<subject_label>_task-<task_label>_atlas-<atlas_label>[_res-<scale_label>]_timeseries.matwhere:
<atlas_label>:Desikan/L2018is the parcellation scheme used<scale_label>:scale1,scale2,scale3,scale4,scale5corresponds to the parcellation scale if applicable
The functional frequency- and time-frequency-domain based connectivity graphs:
eeg/sub-<subject_label>_task-<task_label>_atlas-<atlas_label>[_res-<scale_label>]_conndata-network_connectivity.<fmt>where:
<atlas_label>:Desikan/L2018is the parcellation scheme used<scale_label>:scale1,scale2,scale3,scale4,scale5corresponds to the parcellation scale if applicable<fmt>:mat/gpickle/tsv/graphmlis the format used to store the graph
FreeSurfer Derivatives
A FreeSurfer subjects directory is created in <bids_dataset/derivatives>/freesurfer-7.2.0.
freesurfer-7.1.1/
fsaverage/
mri/
surf/
...
sub-<subject_label>/
mri/
surf/
...
...
The fsaverage subject distributed with the running version of FreeSurfer is copied into this directory.
Nipype Workflow Derivatives
The execution of each Nipype workflow (pipeline) dedicated to the processing of one modality (i.e. anatomical/diffusion/fMRI/EEG) involves the creation of a number of intermediate outputs which are written to <bids_dataset/derivatives>/nipype/sub-<subject_label>/<anatomical/diffusion/fMRI/eeg>_pipeline respectively:

To enhance transparency on how data is processed, outputs include a pipeline execution graph saved as <anatomical/diffusion/fMRI/eeg>_pipeline/graph.svg which summarizes all processing nodes involves in the given processing pipeline:
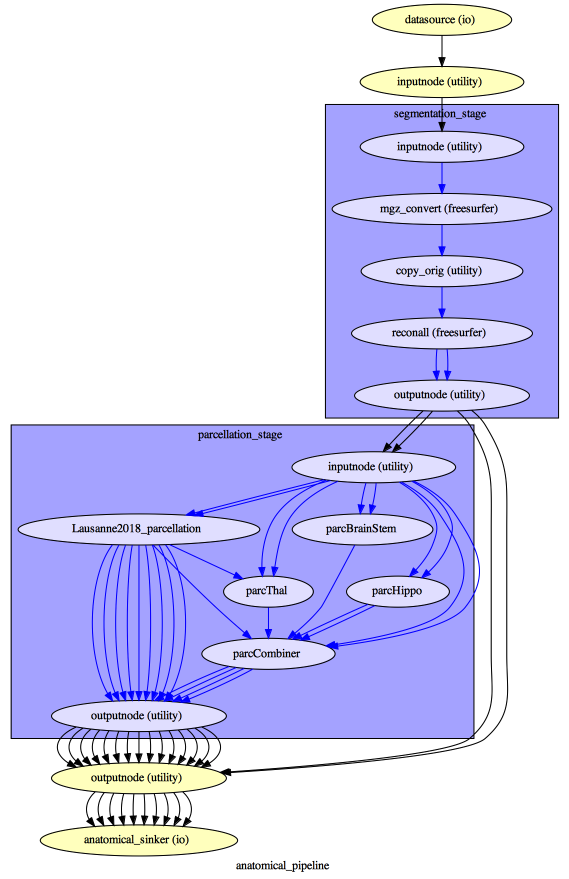
Execution details (data provenance) of each interface (node) of a given pipeline are reported in <anatomical/diffusion/fMRI/eeg>_pipeline/<stage_name>/<interface_name>/_report/report.rst
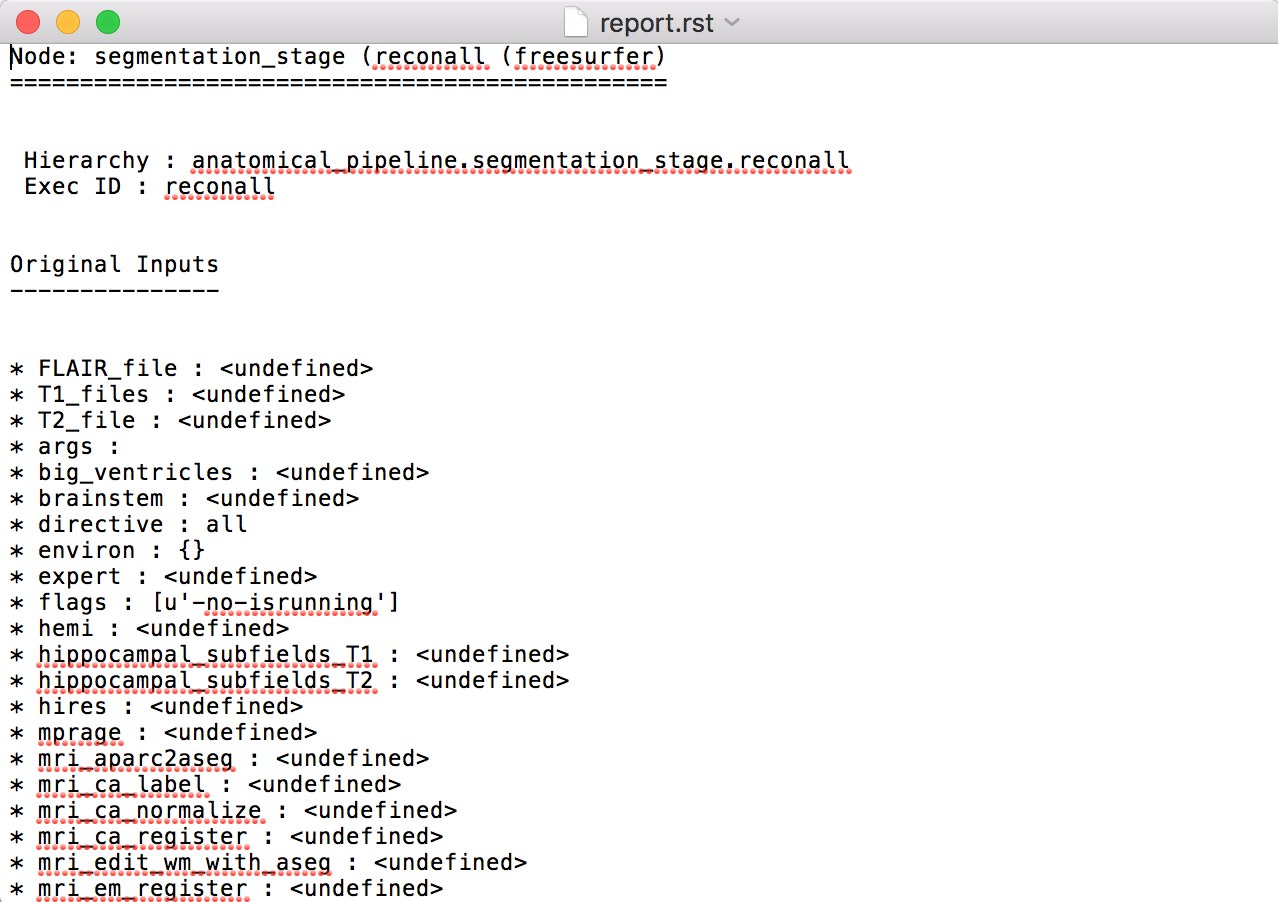
Note
Connectome Mapper 3 outputs are currently being updated to conform to BIDS v1.4.0.